Carry out the following steps:
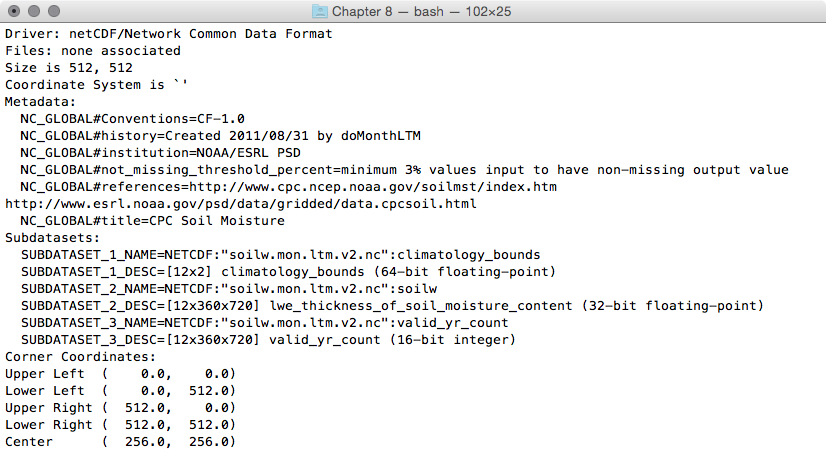
- As the first step, investigate the NetCDF format of the dataset you downloaded using gdalinfo. This kind of dataset is composed of several subdatasets, as you may have realized by looking at the gdalinfo output:
$ gdalinfo NETCDF:"soilw.mon.ltm.v2.nc"
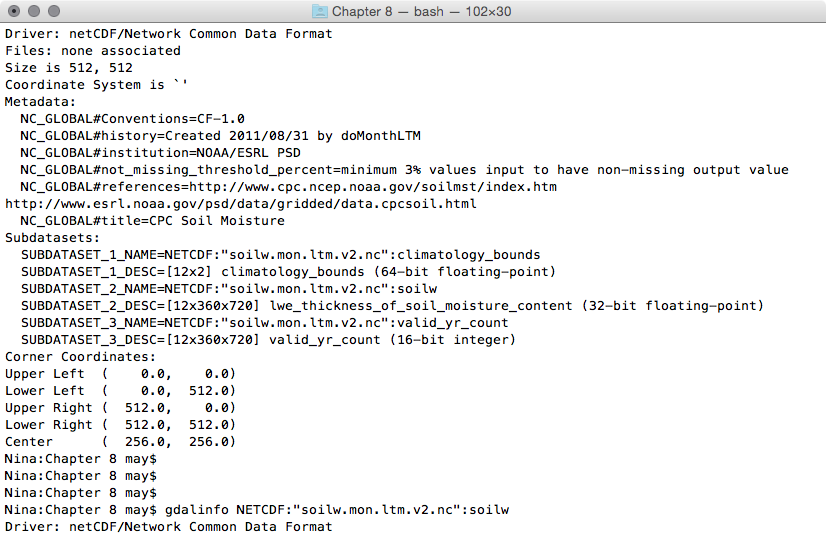
- Use gdalinfo to investigate one of the file's subdatasets. The syntax that the NetCDF GDAL driver (http://www.gdal.org/frmt_netcdf.html) uses is to append a colon followed by the variable name at the end of the filename. For example, try to figure out how many bands the soilw subdataset is composed of. This subdataset, representing lwe_thickness_of_soil_moisture_content, is composed of 12 bands. Each band, according to the information derived by its metadata, represents the CPC Monthly Soil Moisture for a given month. The month is identified by the NETCDF_DIM_time metadata value, which is the number of days from the beginning of the year (0 for January, 31 for February, 59 for March, and so on):
$ gdalinfo NETCDF:"soilw.mon.ltm.v2.nc":soilw
 '
'...(12 bands)...
- What you are going to do is create a Python script using GDAL and NumPy. You will read a given NetCDF dataset, iterate its subdatasets, and then iterate each subdataset's bands. For each subdataset, you will create a point PostGIS layer, and you will add a field for each band in order to store the band values in the layer table. Then, you will iterate the band's cells, and for each cell, you will add a point in the layer with the corresponding band's values. Therefore, create a netcdf2postgis.py file and add the following Python code to it:
import sys
from osgeo import gdal, ogr, osr
from osgeo.gdalconst import GA_ReadOnly, GA_Update
def netcdf2postgis(file_nc, pg_connection_string,
postgis_table_prefix):
# register gdal drivers
gdal.AllRegister()
# postgis driver, needed to create the tables
driver = ogr.GetDriverByName('PostgreSQL')
srs = osr.SpatialReference()
# for simplicity we will assume all of the bands in the datasets
# are in the same spatial reference, wgs 84
srs.ImportFromEPSG(4326)
# first, check if dataset exists
ds = gdal.Open(file_nc, GA_ReadOnly)
if ds is None:
print 'Cannot open ' + file_nc
sys.exit(1)
# 1. iterate subdatasets
for sds in ds.GetSubDatasets():
dataset_name = sds[0]
variable = sds[0].split(':')[-1]
print 'Importing from %s the variable %s...' %
(dataset_name, variable)
# open subdataset and read its properties
sds = gdal.Open(dataset_name, GA_ReadOnly)
cols = sds.RasterXSize
rows = sds.RasterYSize
bands = sds.RasterCount
# create a PostGIS table for the subdataset variable
table_name = '%s_%s' % (postgis_table_prefix, variable)
pg_ds = ogr.Open(pg_connection_string, GA_Update )
pg_layer = pg_ds.CreateLayer(table_name, srs = srs,
geom_type=ogr.wkbPoint, options = [
'GEOMETRY_NAME=the_geom',
'OVERWRITE=YES',
# this will drop and recreate the table every time
'SCHEMA=chp08',
])
print 'Table %s created.' % table_name
# get georeference transformation information
transform = sds.GetGeoTransform()
pixelWidth = transform[1]
pixelHeight = transform[5]
xOrigin = transform[0] + (pixelWidth/2)
yOrigin = transform[3] - (pixelWidth/2)
# 2. iterate subdataset bands and append them to data
data = []
for b in range(1, bands+1):
band = sds.GetRasterBand(b)
band_data = band.ReadAsArray(0, 0, cols, rows)
data.append(band_data)
# here we add the fields to the table, a field for each band
# check datatype (Float32, 'Float64', ...)
datatype = gdal.GetDataTypeName(band.DataType)
ogr_ft = ogr.OFTString # default for a field is string
if datatype in ('Float32', 'Float64'):
ogr_ft = ogr.OFTReal
elif datatype in ('Int16', 'Int32'):
ogr_ft = ogr.OFTInteger
# here we add the field to the PostGIS layer
fd_band = ogr.FieldDefn('band_%s' % b, ogr_ft)
pg_layer.CreateField(fd_band)
print 'Field band_%s created.' % b
# 3. iterate rows and cols
for r in range(0, rows):
y = yOrigin + (r * pixelHeight)
for c in range(0, cols):
x = xOrigin + (c * pixelWidth)
# for each cell, let's add a point feature
# in the PostGIS table
point_wkt = 'POINT(%s %s)' % (x, y)
point = ogr.CreateGeometryFromWkt(point_wkt)
featureDefn = pg_layer.GetLayerDefn()
feature = ogr.Feature(featureDefn)
# now iterate bands, and add a value for each table's field
for b in range(1, bands+1):
band = sds.GetRasterBand(1)
datatype = gdal.GetDataTypeName(band.DataType)
value = data[b-1][r,c]
print 'Storing a value for variable %s in point x: %s,
y: %s, band: %s, value: %s' % (variable, x, y, b, value)
if datatype in ('Float32', 'Float64'):
value = float(data[b-1][r,c])
elif datatype in ('Int16', 'Int32'):
value = int(data[b-1][r,c])
else:
value = data[r,c]
feature.SetField('band_%s' % b, value)
# set the feature's geometry and finalize its creation
feature.SetGeometry(point) pg_layer.CreateFeature(feature)
- To run the netcdf2postgis method from the command line, add the entry point for the script. The code will check whether the script user is correctly using the three required parameters, which are the NetCDF file path, the GDAL PostGIS connection string, and a prefix/suffix to use for table names in PostGIS:
if __name__ == '__main__':
# the user must provide at least three parameters,
# the netCDF file path, the PostGIS GDAL connection string # and the prefix suffix to use for PostGIS table names
if len(sys.argv) < 4 or len(sys.argv) > 4:
print "usage: <netCDF file path> <GDAL PostGIS connection
string><PostGIS table prefix>"
raise SystemExit
file_nc = sys.argv[1]
pg_connection_string = sys.argv[2]
postgis_table_prefix = sys.argv[3] netcdf2postgis(file_nc, pg_connection_string,
postgis_table_prefix)
- Run the script. Be sure to use the correct NetCDF file path, GDAL PostGIS connection string (check the format at http://www.gdal.org/drv_pg.html), and a table prefix that has to be appended to the table names for tables that will be created in PostGIS:
(postgis-cb-env)$ python netcdf2postgis.py
NETCDF:"soilw.mon.ltm.v2.nc"
"PG:dbname='postgis_cookbook' host='localhost'
port='5432' user='me' password='mypassword'" netcdf
Importing from NETCDF:"soilw.mon.ltm.v2.nc":
climatology_bounds the variable climatology_bounds...
...
Importing from NETCDF:"soilw.mon.ltm.v2.nc":soilw the
variable soilw...
Table netcdf_soilw created.
Field band_1 created.
Field band_2 created.
...
Field band_11 created.
Field band_12 created.
Storing a value for variable soilw in point x: 0.25,
y: 89.75, band: 2, value: -9.96921e+36
Storing a value for variable soilw in point x: 0.25,
y: 89.75, band: 3, value: -9.96921e+36 ...
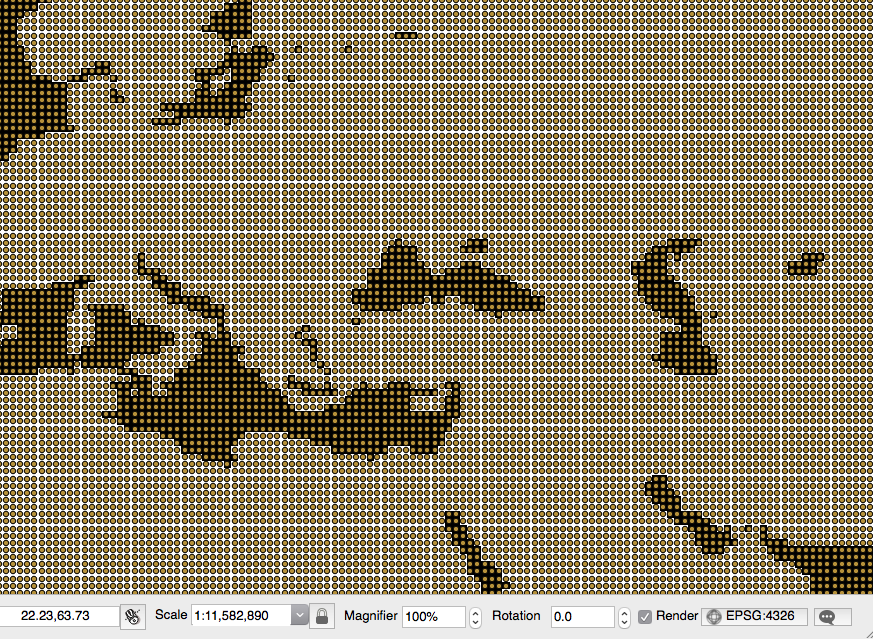
- At the end of the process, check the results by opening one of the output PostGIS tables using your favorite GIS desktop tool. The following screenshot shows how it looks in the QGIS soilw layer with the original NetCDF dataset behind it: