Chapter 10. Graphics
Introduction
Graphics is a great strength of R. The graphics package is part of the
standard distribution and contains many useful functions for creating a
variety of graphic displays. The base functionality has been expanded
and made easier with ggplot2, part of the tidyverse of packages. In
this chapter we will focus on examples using ggplot2, and we will
occasionally suggest other packages. In this chapter’s See Also sections
we mention functions in other packages that do the same job in a
different way. We suggest that you explore those alternatives if you are
dissatisfied with what’s offered by ggplot2 or base graphics.
Graphics is a vast subject, and we can only scratch the surface here.
Winston Chang’s R Graphics Cookbook, 2nd Edition is part of the
O’Reilly Cookbook series and walks through many useful recipes with a
focus on ggplot2. If you want to delve deeper, we recommend R
Graphics by Paul Murrell (Chapman & Hall, 2006). That book discusses
the paradigms behind R graphics, explains how to use the graphics
functions, and contains numerous examples—including the code to recreate
them. Some of the examples are pretty amazing.
The Illustrations
The graphs in this chapter are mostly plain and unadorned. We did that
intentionally. When you call the ggplot function, as in:
library(tidyverse)
Figure 10-1. Simple Plot
df<-data.frame(x=1:5,y=1:5)ggplot(df,aes(x,y))+geom_point()
you get a plain, graphical representation of x and y as shown in
Figure 10-1. You could adorn the graph with colors, a
title, labels, a legend, text, and so forth, but then the call to
ggplot becomes more and more crowded, obscuring the basic intention.
ggplot(df,aes(x,y))+geom_point()+labs(title="Simple Plot Example",subtitle="with a subtitle",x="x values",y="y values")+theme(panel.background=element_rect(fill="white",colour="grey50"))
Figure 10-2. Complicated Plot
The resulting plot is shown in Figure 10-2. We want to keep the recipes clean, so we emphasize the basic plot and then show later (as in “Adding a Title and Labels”) how to add adornments.
Notes on ggplot2 basics
While the package is called ggplot2 the primary plotting function in
the package is called ggplot. It is important to understand the basic
pieces of a ggplot2 graph. In the examples above you can see that we
pass data into ggplot then define how the graph is created by stacking
together small phrases that describe some aspect of the plot. This
stacking together of phrases is part of the “grammar of graphics” ethos
(that’s where the gg comes from). To learn more, you can read “The
Layered Grammar of Graphics” written by ggplot2 author, Hadley Wickham
(http://vita.had.co.nz/papers/layered-grammar.pdf). The “grammar of
graphics,” concept originating with Leland Wilkinson who articulated the
idea of building graphics up from a set of primitives (i.e. verbs and
nouns). With ggplot, the underlying data need not be fundamentally
reshaped for each type of graphical representation. In general, the data
stays the same and the user then changes syntax slightly to illustrate
the data differently. This is significantly more consistent than base
graphics which often require reshaping the data in order to change the
way the data is visualized.
As we talk about ggplot graphics it’s worth defining the things that
make up a ggplot graph:
geometric object functions-
These are geometric objects that describe the type of graph being created. These start with
geom_and examples includegeom_line,geom_boxplot, andgeom_pointalong with dozens more. aesthetics-
The aesthetics, or aesthetic mappings, communicate to
ggplotwhich fields in the source data get mapped to which visual elements in the graphic. This is theaes()line in aggplotcall. stats-
Stats are statistical transformations that are done before displaying the data. Not all graphs will have stats, but a few common stats are
stat_ecdf(the empirical cumulative distribution function) andstat_identitywhich tellsggplotto pass the data without doing any stats at all. facet functions-
Facets are subplots where each small plot represents a subgroup of the data. The faceting functions include
facet_wrapandfacet_grid. themes-
Themes are the visual elements of the plot that are not tied to data. These might include titles, margins, table of contents locations, or font choices.
layer-
A layer is a combination of data, aesthetics, a geometric object, a stat, and other options to produce a visual layer in the
ggplotgraphic.
“Long” vs. “Wide” data with ggplot
One of the first confusions new users to ggplot often face is that
they are inclined to reshape their data to be “wide” before plotting it.
Wide here meaning every variable they are plotting is its own column in
the underlying data frame.
ggplot works most easily with “long” data where additional variables
are added as rows in the data frame rather than columns. The great side
effect of adding additional measurements as rows is that any properly
constructed ggplot graphs will automatically update to reflect the new
data without changing the ggplot code. If each additional variable was
added as a column, then the plotting code would have to be changed to
introduce additional variables. This idea of “long” vs. “wide” data will
become more obvious in the examples in the rest of this chapter.
Graphics in Other Packages
R is highly programmable, and many people have extended its graphics
machinery with additional features. Quite often, packages include
specialized functions for plotting their results and objects. The zoo
package, for example, implements a time series object. If you create a
zoo object z and call plot(z), then the zoo package does the
plotting; it creates a graphic that is customized for displaying a time
series. Zoo uses base graphics so the resulting graph will not be a
ggplot graphic.
There are even entire packages devoted to extending R with new graphics
paradigms. The lattice package is an alternative to base graphics that
predates ggplot2. It uses a powerful graphics paradigm that enables
you to create informative graphics more easily. It was implemented by
Deepayan Sarkar, who also wrote Lattice: Multivariate Data
Visualization with R (Springer, 2008), which explains the package and
how to use it. The lattice package is also described in
R in a Nutshell (O’Reilly).
There are two chapters in Hadley Wickham’s excellent book R for Data
Science which deal with graphics. The first, “Exploratory Data
Analysis” focuses on exploring data with ggplot2 while “Graphics for
Communication” explores communicating to others with graphics. R for
Data Science is availiable in a printed version from O’Reilly Media or
online at http://r4ds.had.co.nz/graphics-for-communication.html.
Creating a Scatter Plot
Problem
You have paired observations: (x1, y1), (x2, y2), …, (xn, yn). You want to create a scatter plot of the pairs.
Solution
We can plot the data by calling ggplot, passing in the data frame, and
invoking a geometric point function:
ggplot(df,aes(x,y))+geom_point()
In this example, the data frame is called df and the x and y data
are in fields named x and y which we pass to the aesthetic in the
call aes(x, y).
Discussion
A scatter plot is a common first attack on a new dataset. It’s a quick way to see the relationship, if any, between x and y.
Plotting with ggplot requires telling ggplot what data frame to use,
then what type of graph to create, and which aesthetic mapping (aes) to
use . The aes in this case defines which field from df goes into
which axis on the plot. Then the command geom_point communicates that
you want a point graph, as opposed to a line or other type of graphic.
We can use the built in mtcars dataset to illustrate plotting
horsepower hp on the x axis and fuel economy mpg on the y:
ggplot(mtcars,aes(hp,mpg))+geom_point()
Figure 10-3. Scatter Plot Example
The resulting plot is shown in Figure 10-3.
See Also
See “Adding a Title and Labels” for adding a title and labels; see Recipes and for adding a grid and a legend (respectively). See “Plotting All Variables Against All Other Variables” for plotting multiple variables.
Adding a Title and Labels
Problem
You want to add a title to your plot or add labels for the axes.
Solution
With ggplot we add a labs element which controls the labels for the
title and axies.
When calling labs in ggplot:
: title The desired title text
: x x-axis label
: y: y-axis label
ggplot(df,aes(x,y))+geom_point()+labs(title="The Title",x="X-axis Label",y="Y-axis Label")
Discussion
The graph created in “Creating a Scatter Plot” is quite plain. A title and better labels will make it more interesting and easier to interpret.
Note that in ggplot you build up the elements of the graph by
connecting the parts with the plus sign +. So we add additional
graphical elements by stringing together phrases. You can see this in
the following code that uses the built in cars dataset and plots speed
vs. stopping distance in a scatter plot, shown in Figure 10-4
ggplot(mtcars,aes(hp,mpg))+geom_point()+labs(title="Cars: Horsepower vs. Fuel Economy",x="HP",y="Economy (miles per gallon)")
Figure 10-4. Labeled Axis and Title
Adding (or Removing) a Grid
Problem
You want to change the background grid to your graphic.
Solution
With ggplot background grids come as a default, as you have seen in
other recipes. However we can alter the background grid using the
theme function or by applying a prepackaed theme to our graph.
We can use theme to alter the backgorund panel of our graphic:
ggplot(df)+geom_point(aes(x,y))+theme(panel.background=element_rect(fill="white",colour="grey50"))
Figure 10-5. White background
Discussion
ggplot fills in the background with a grey grid by default. So you may
find yourself wanting to remove that grid completely or change it to
something else. Let’s create a ggplot graphic and then incrementally
change the background style.
We can add or change aspects of our graphic by creating a ggplot
object then calling the object and using the + to add to it. The
background shading in a ggplot graphic is actually 3 different graph
elements:
panel.grid.major:
These are white by default and heavy
panel.grid.minor:
These are white by default and light
panel.background:
This is the background that is grey by default
You can see these elements if you look carefully at the background of Figure 10-4:
If we set the background as element_blank() then the major and minor
grids are there but they are white on white so we can’t see them in
???:
g1<-ggplot(mtcars,aes(hp,mpg))+geom_point()+labs(title="Cars: Horsepower vs. Fuel Economy",x="HP",y="Economy (miles per gallon)")+theme(panel.background=element_blank())g1
. image::images/10_Graphics_files/figure-html/examplebackground-1.png[]
Notice in the code above we put the ggplot graph into a variable
called g1. Then we printed the graphic by just calling g1. By having
the graph inside of g1 we can then add additional graphical components
without rebuilding the graph again.
But if we wanted to show the background grid in some bright colors for illustration, it’s as easy as setting them to a color and setting a line type which is shown in ???.
g2<-g1+theme(panel.grid.major=element_line(color="red",linetype=3))+# linetype = 3 is dashtheme(panel.grid.minor=element_line(color="blue",linetype=4))# linetype = 4 is dot dashg2
. image::images/10_Graphics_files/figure-html/majorgrid-1.png[]
??? lacks visual appeal, but you can cleary see that the red lines make up the major grid and the blue lines are the minor grid.
Or we could do something less garash and take the ggplot object g1
from above and add grey gridlines to the white background, shown in
Figure 10-6.
g1+theme(panel.grid.major=element_line(colour="grey"))
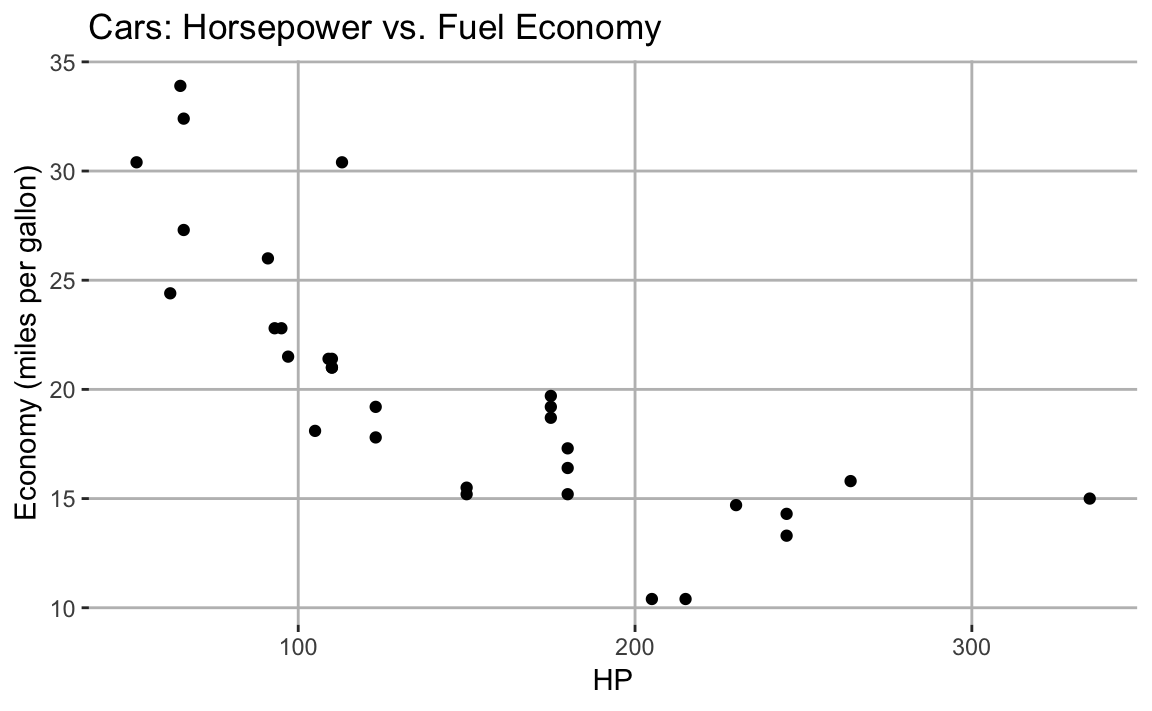
Figure 10-6. Grey Major Gridlines
Creating a Scatter Plot of Multiple Groups
Problem
You have data in a data frame with three observations per record: x, y, and a factor f that indicates the group. You want to create a scatter plot of x and y that distinguishes among the groups.
Solution
With ggplot we control the mapping of shapes to the factor f by
passing shape = f to the aes.
ggplot(df,aes(x,y,shape=f))+geom_point()
Discussion
Plotting multiple groups in one scatter plot creates an uninformative
mess unless we distinguish one group from another. This distinction is
done in ggplot by setting the shape parameter of the aes function.
The built in iris dataset contains paired measures of Petal.Length
and Petal.Width. Each measurement also has a Species property
indicating the species of the flower that was measured. If we plot all
the data at once, we just get the scatter plot shown in ???:
ggplot(data=iris,aes(x=Petal.Length,y=Petal.Width))+geom_point()
. image::images/10_Graphics_files/figure-html/irisnoshape-1.png[]
The graphic would be far more informative if we distinguished the points
by species. In addition to distinguising species by shape, we could also
differentiate by color. We can add shape = Species and
color = Species to our aes call, to get each species with a
different shape and color, shown in ???.
ggplot(data=iris,aes(x=Petal.Length,y=Petal.Width,shape=Species,color=Species))+geom_point()
. image::images/10_Graphics_files/figure-html/irisshape-1.png[]
ggplot conveniently sets up a legend for you as well, which is handy.
See Also
See “Adding (or Removing) a Legend” to add a legend.
Adding (or Removing) a Legend
Problem
You want your plot to include a legend, the little box that decodes the graphic for the viewer.
Solution
In most cases ggplot will add the legends automatically, as you can
see in the previous recipe. If you do not have explicit grouping in the
aes then ggplot will not show a legend by default. If we want to
force ggplot to show a legend we can set the shape or linetype of our
graph to a constant. ggplot will then show a legend with one group. We
then use guides to guide ggplot in how to label the legend.
This can be illustrated with our iris scatterplot:
g<-ggplot(data=iris,aes(x=Petal.Length,y=Petal.Width,shape="Point Name"))+geom_point()+guides(shape=guide_legend(title="Legend Title"))g
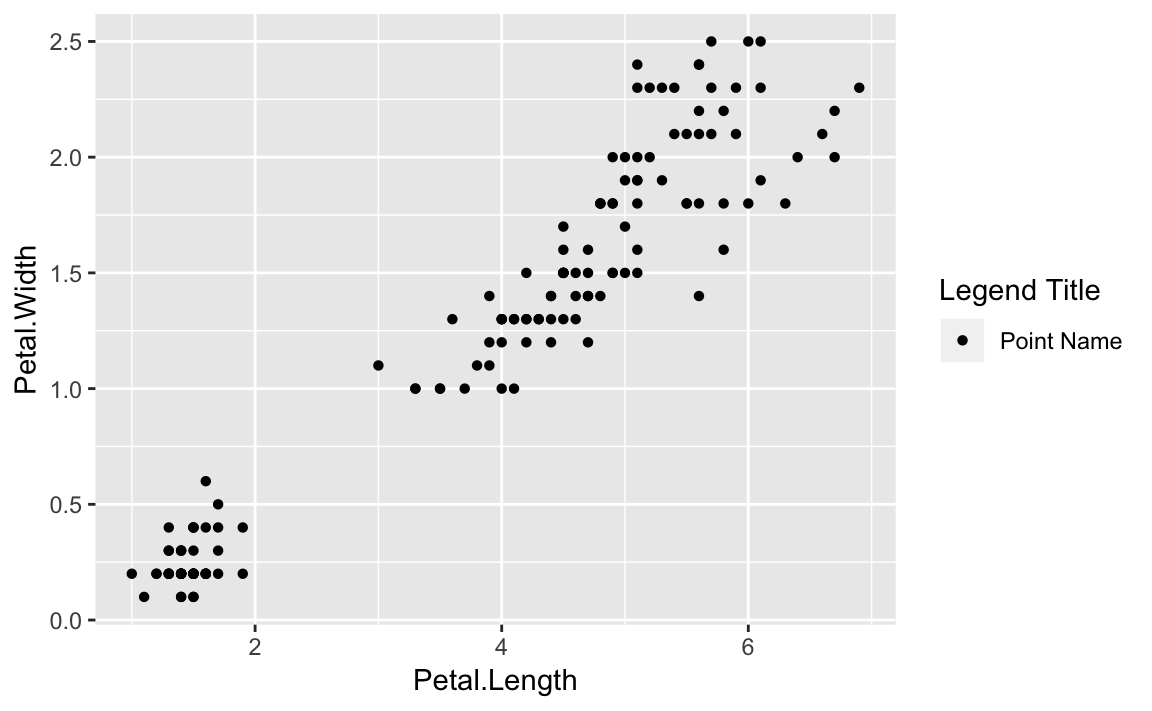
Figure 10-7. Legend Added
Figure 10-7 illustrates the result of setting the shape
to a string value then relabeling the legend using guides.
More commonly, you may want to turn legends off which can be done by
setting the legend.position = "none" in the theme. We can use the
iris plot from the prior recipe and add the theme call as shown in
Figure 10-8:
g<-ggplot(data=iris,aes(x=Petal.Length,y=Petal.Width,shape=Species,color=Species))+geom_point()+theme(legend.position="none")g
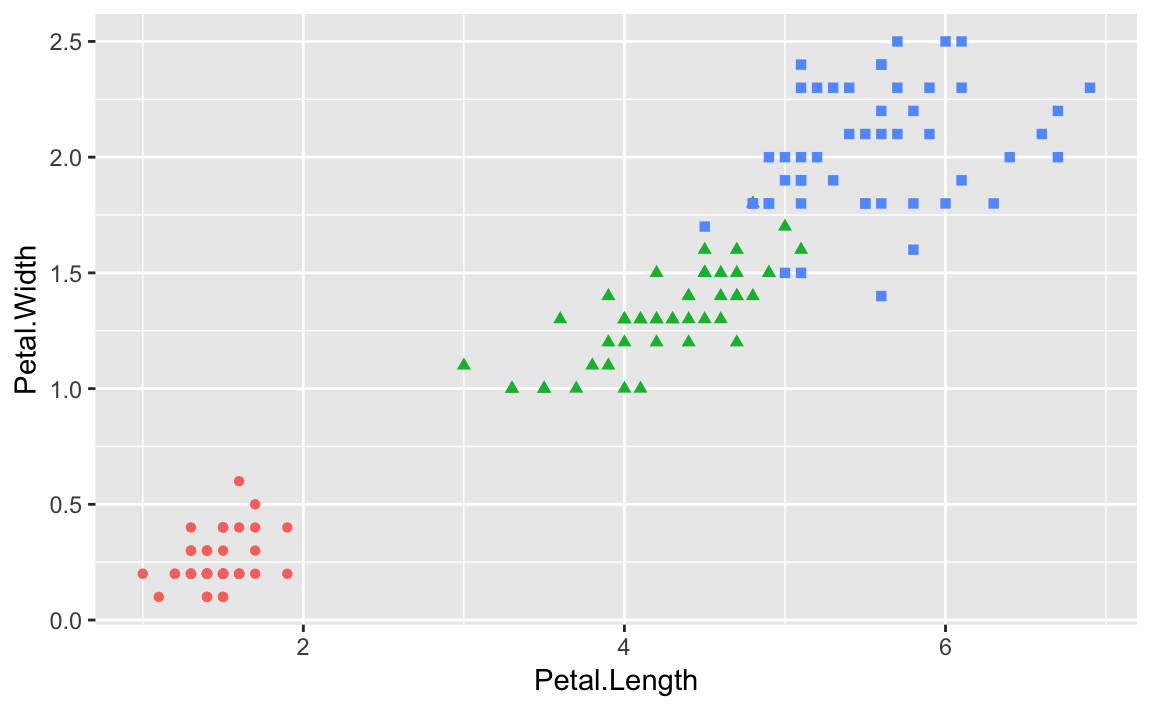
Figure 10-8. Legend Removed
Discussion
Adding legends to ggplot when there is no grouping is an excercise in
tricking ggplot into showing the legend by passing a string to a
grouping parameter in aes. This will not change the grouping as there
is only one group, but will result in a legend being shown with a name.
Then we can use guides to alter the legend title. It’s worth noting
that we are not changing anything about the data, just exploiting
settings in order to coerce ggplot into showing a legend when it
typically would not.
One of the huge benefits of ggplot is its very good defaults. Getting
positions and correspondence between labels and their point types is
done automatically, but can be overridden if needed. To remove a legend
totally, we set theme parameters with
theme(legend.position = "none"). In addition to “none” you can set the
legend.position to be "left", "right", "bottom", "top", or a
two-element numeric vector. Use a two-element numeric vector in order to
pass ggplot specific coordinates of where you want the legend. If
using the coordinate positions the values passed are between 0 and 1 for
x and y position, respectivly.
An example of a legend positioned at the bottom is in Figure 10-9 created with this adjustment to the
legend.poisition:
g+theme(legend.position="bottom")
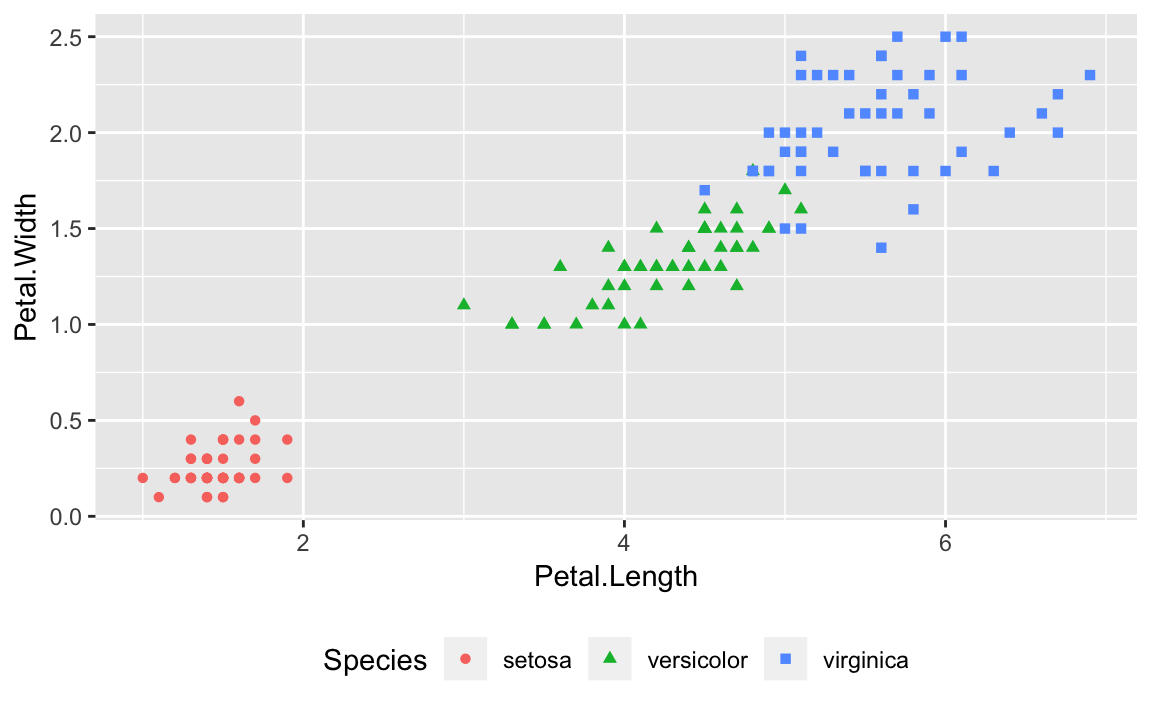
Figure 10-9. Legend on the Bottom
Or we could use the two-element numeric vector to put the legend in a specific location as in Figure 10-10. The example puts the center of the legend at 80% to the right and 20% up from the bottom.
g+theme(legend.position=c(.8,.2))
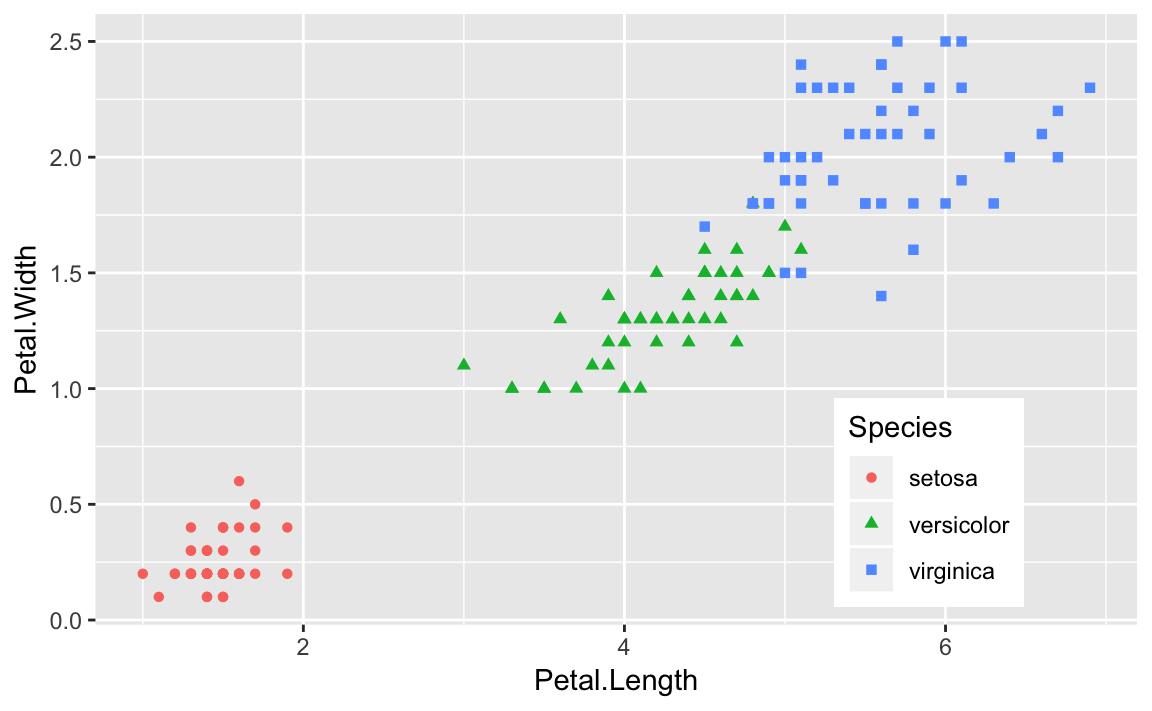
Figure 10-10. Legend at a Point
In many aspects beyond legends, ggplot uses sane defaults with
flexibility to override those and tweak the details. More detail of
ggplot options related to legends can be found in the help for theme
by typing ?theme or looking in the ggplot online reference material:
Plotting the Regression Line of a Scatter Plot
Problem
You are plotting pairs of data points, and you want to add a line that illustrates their linear regression.
Solution
Using ggplot there is no need to calculate the linear model first
using the R lm function. We can instead use the geom_smooth function
to calculate the linear regression inside of our ggplot call.
If our data is in a data frame df and the x and y data are in
columns x and y we plot the regression line like this:
ggplot(df,aes(x,y))+geom_point()+geom_smooth(method="lm",formula=y~x,se=FALSE)
The se = FALSE parameter tells ggplot not to plot the standard error
bands around our regression line.
Discussion
Suppose we are modeling the strongx dataset found in the faraway
package. We can create a linear model using the built in lm function
in R. We can predict the variable crossx as a linear function of
energy. First, lets look at a simple scatter plot of our data:
library(faraway)data(strongx)ggplot(strongx,aes(energy,crossx))+geom_point()
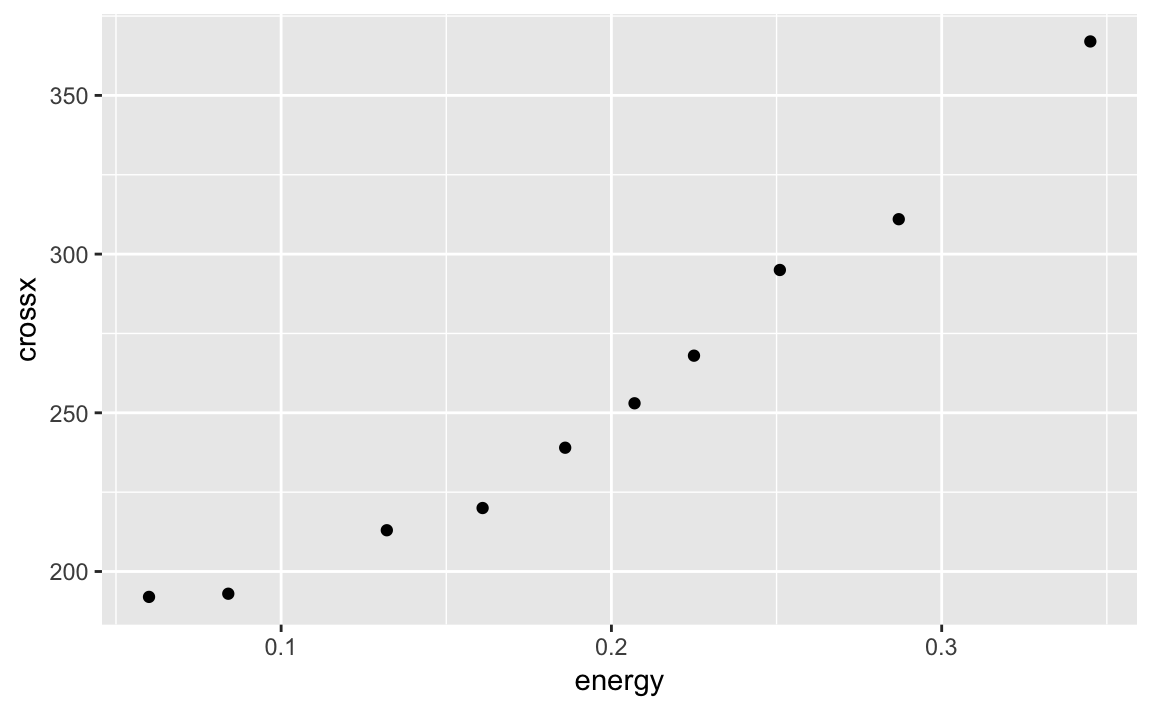
Figure 10-11. Strongx Scatter Plot
ggplot can calculate a linear model on the fly and then plot the
regression line along with our data:
g<-ggplot(strongx,aes(energy,crossx))+geom_point()g+geom_smooth(method="lm",formula=y~x,se=FALSE)
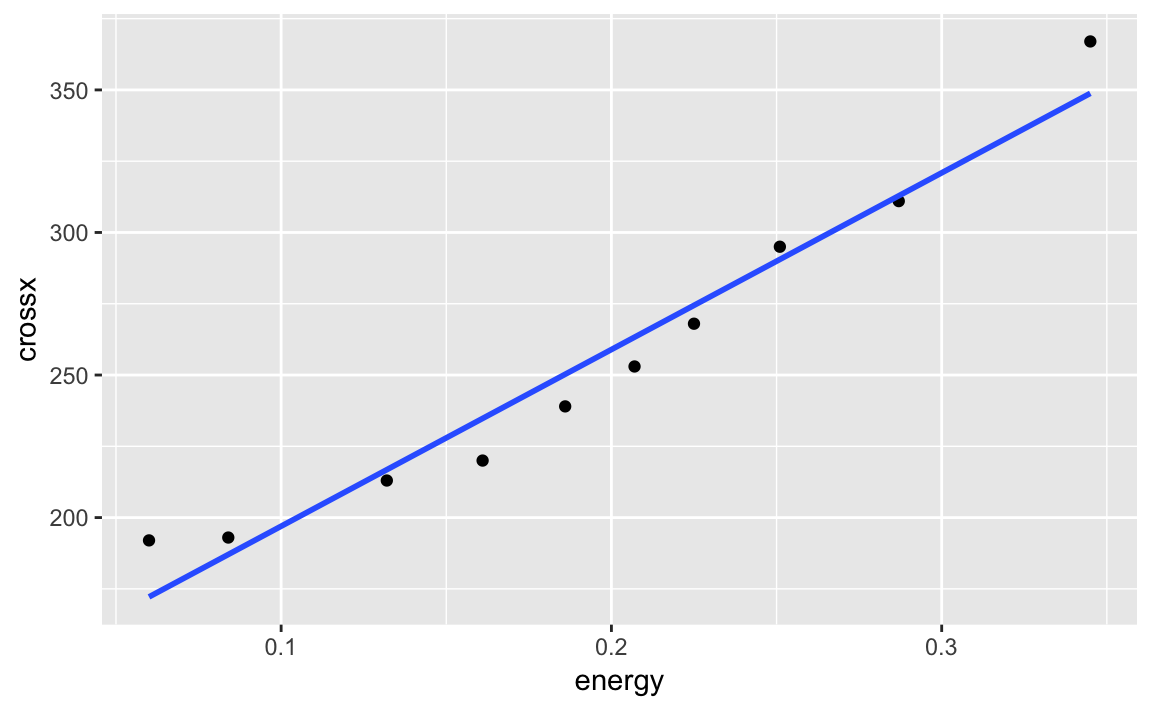
We can turn the confidence bands on by omitting the se = FALSE option
as as shown in ???:
g+geom_smooth(method="lm",formula=y~x)
. image::images/10_Graphics_files/figure-html/one-step-nose-1.png[]
Notice that in the geom_smooth we use x and y rather than the
variable names. ggplot has set the x and y inside the plot based
on the aesthetic. Multiple smoothing methods are supported by
geom_smooth. You can explore those, and other options in the help by
typing ?geom_smooth.
If we had a line we wanted to plot that was stored in another R object,
we could use geom_abline to plot the line on our graph. In the
following example we pull the intercept term and the slope from the
regression model m and add those to our graph:
m<-lm(crossx~energy,data=strongx)ggplot(strongx,aes(energy,crossx))+geom_point()+geom_abline(intercept=m$coefficients[1],slope=m$coefficients[2])
This produces a very similar plot to ???. The
geom_abline method can be handy if you are plotting a line from a
source other than a simple linear model.
See Also
See the chapter on Linear Regression and
ANOVA for more about linear regression and the lm function.
Plotting All Variables Against All Other Variables
Problem
Your dataset contains multiple numeric variables. You want to see scatter plots for all pairs of variables.
Solution
ggplot does not have any built in method to create pairs plots,
however, the package GGally provides the functionality with the
ggpairs function:
library(GGally)ggpairs(df)
Discussion
When you have a large number of variables, finding interrelationships
between them is difficult. One useful technique is looking at scatter
plots of all pairs of variables. This would be quite tedious if coded
pair-by-pair, but the ggpairs function from the package GGally
provides an easy way to produce all those scatter plots at once.
The iris dataset contains four numeric variables and one categorical
variable:
head(iris)#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species#> 1 5.1 3.5 1.4 0.2 setosa#> 2 4.9 3.0 1.4 0.2 setosa#> 3 4.7 3.2 1.3 0.2 setosa#> 4 4.6 3.1 1.5 0.2 setosa#> 5 5.0 3.6 1.4 0.2 setosa#> 6 5.4 3.9 1.7 0.4 setosa
What is the relationship, if any, between the columns? Plotting the
columns with ggpairs produces multiple scatter plots.
library(GGally)ggpairs(iris)
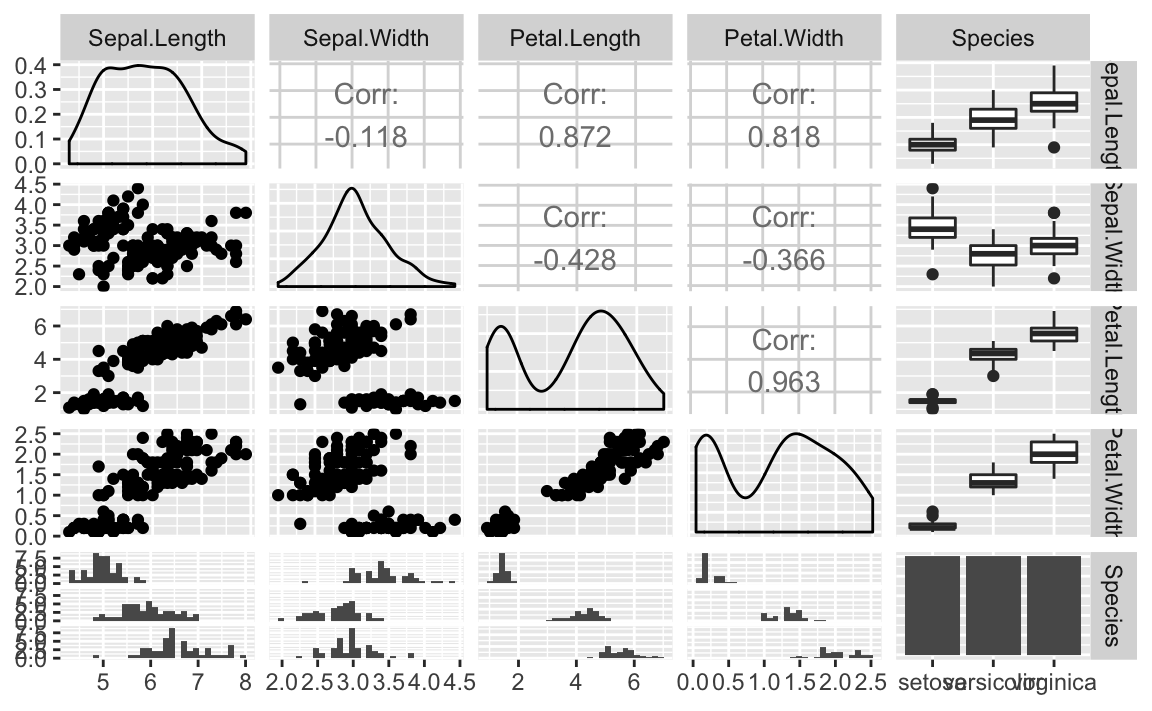
Figure 10-12. ggpairs Plot of Iris Data
The ggpairs function is pretty, but not particularly fast. If you’re
just doing interactive work and want a quick peak at the data, the base
R plot function provides faster output and is shown in Figure 10-13.
plot(iris)
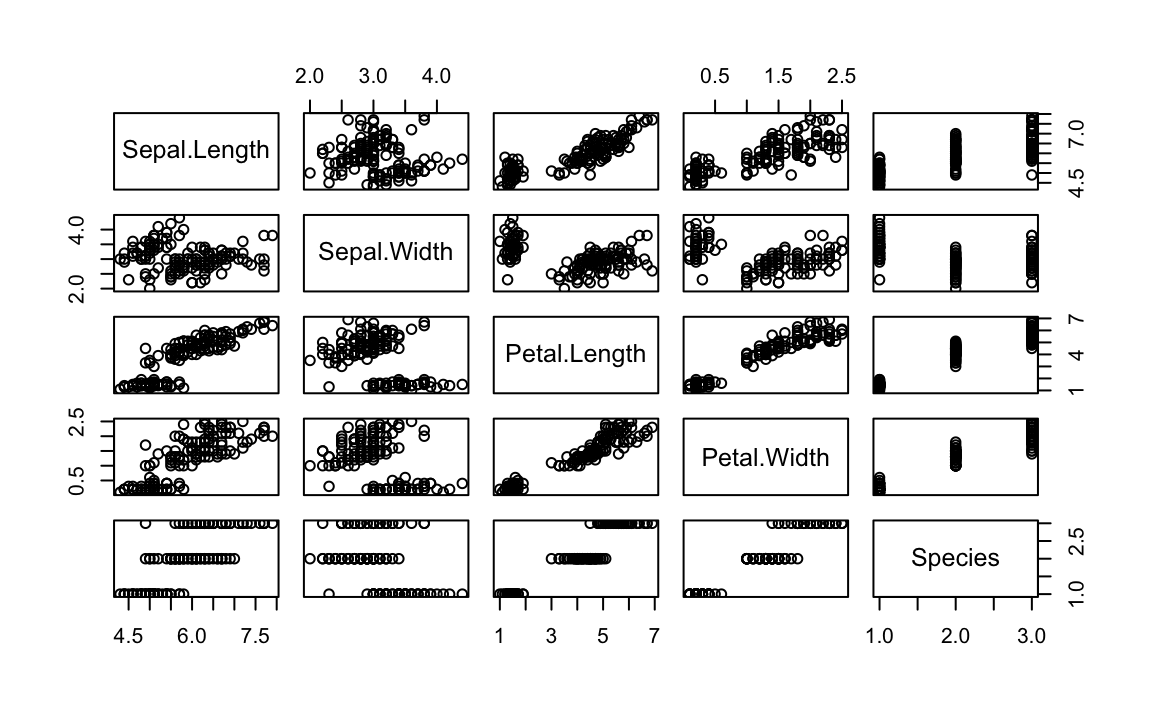
Figure 10-13. Base plot() Pairs Plot
While the ggpairs function is not as fast to plot as the Base R plot
function, it produces density graphs on the diagonal and reports
correlation in the upper triangle of the graph. When factors or
character columns are present, ggpairs produces histograms on the
lower triangle of the graph and boxplots on the upper triangle. These
are nice additions to understanding relationships in your data.
Creating One Scatter Plot for Each Factor Level
Problem
Your dataset contains (at least) two numeric variables and a factor. You want to create several scatter plots for the numeric variables, with one scatter plot for each level of the factor.
Solution
This kind of plot is called a conditioning plot, which is produced in
ggplot by adding facet_wrap to our plot. In this example we use the
data frame df which contains three columns: x, y, and f with f
being a factor (or a character).
ggplot(df,aes(x,y))+geom_point()+facet_wrap(~f)
Discussion
Conditioning plots (coplots) are another way to explore and illustrate the effect of a factor or to compare different groups to each other.
The Cars93 dataset contains 27 variables describing 93 car models as
of 1993. Two numeric variables are MPG.city, the miles per gallon in
the city, and Horsepower, the engine horsepower. One categorical
variable is Origin, which can be USA or non-USA according to where the
model was built.
Exploring the relationship between MPG and horsepower, we might ask: Is there a different relationship for USA models and non-USA models?
Let’s examine this as a facet plot:
data(Cars93,package="MASS")ggplot(data=Cars93,aes(MPG.city,Horsepower))+geom_point()+facet_wrap(~Origin)
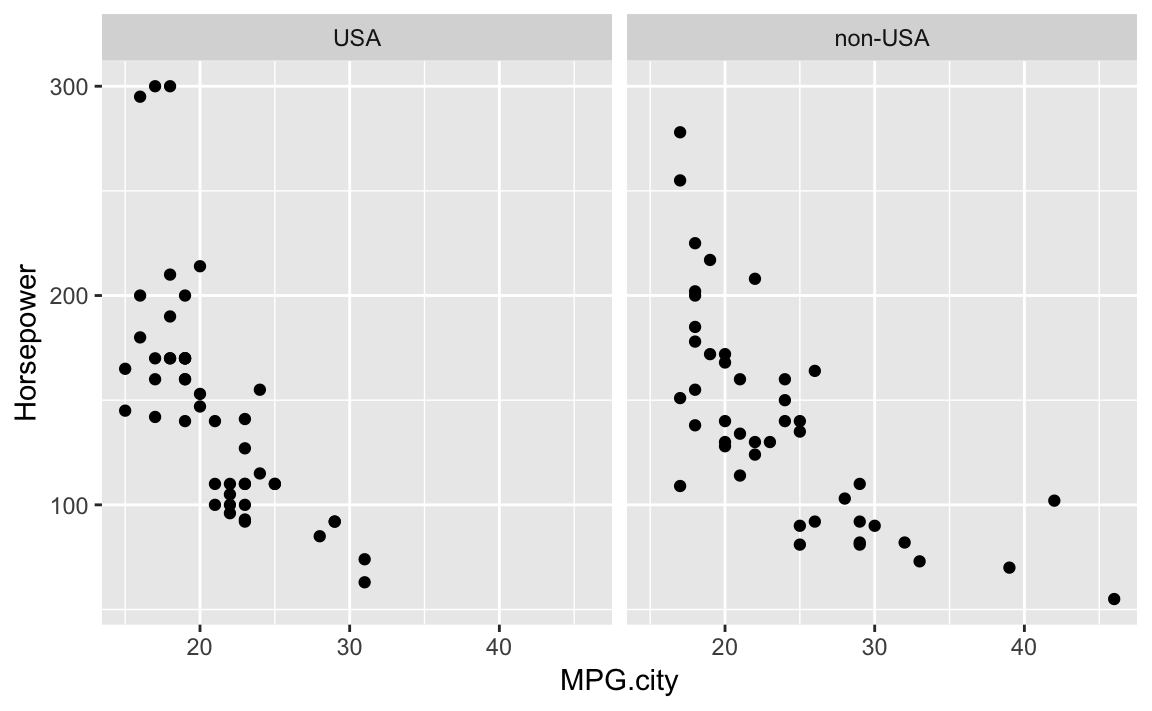
Figure 10-14. Cars Data with Facet
The resulting plot in Figure 10-13 reveals a few insights. If we really crave that 300-horsepower monster then we’ll have to buy a car built in the USA; but if we want high MPG, we have more choices among non-USA models. These insights could be teased out of a statistical analysis, but the visual presentation reveals them much more quickly.
Note that using facet results in subplots with the same x and y
axis ranges. This helps insure that visual inspection of the data is not
misleading because of differeing axis ranges.
See Also
The Base R Graphics function coplot can accomplish very similar plots
using only Base Graphics.
Creating a Bar Chart
Problem
You want to create a bar chart.
Solution
A common situation is to have a column of data that represents a group and then another column that represents a measure about that group. This format is “long” data because the data runs vertically instead of having a column for each group.
Using the geom_bar function in ggplot we can plot the heights as
bars. If the data is already aggregated, we add stat = "identity" so
that ggplot knows it needs to do no aggregation on the groups of
values before plotting.
ggplot(data=df,aes(x,y))+geom_bar(stat="identity")
Discussion
Let’s use the cars made by Ford in the Cars93 data in an example:
ford_cars<-Cars93%>%filter(Manufacturer=="Ford")ggplot(ford_cars,aes(Model,Horsepower))+geom_bar(stat="identity")
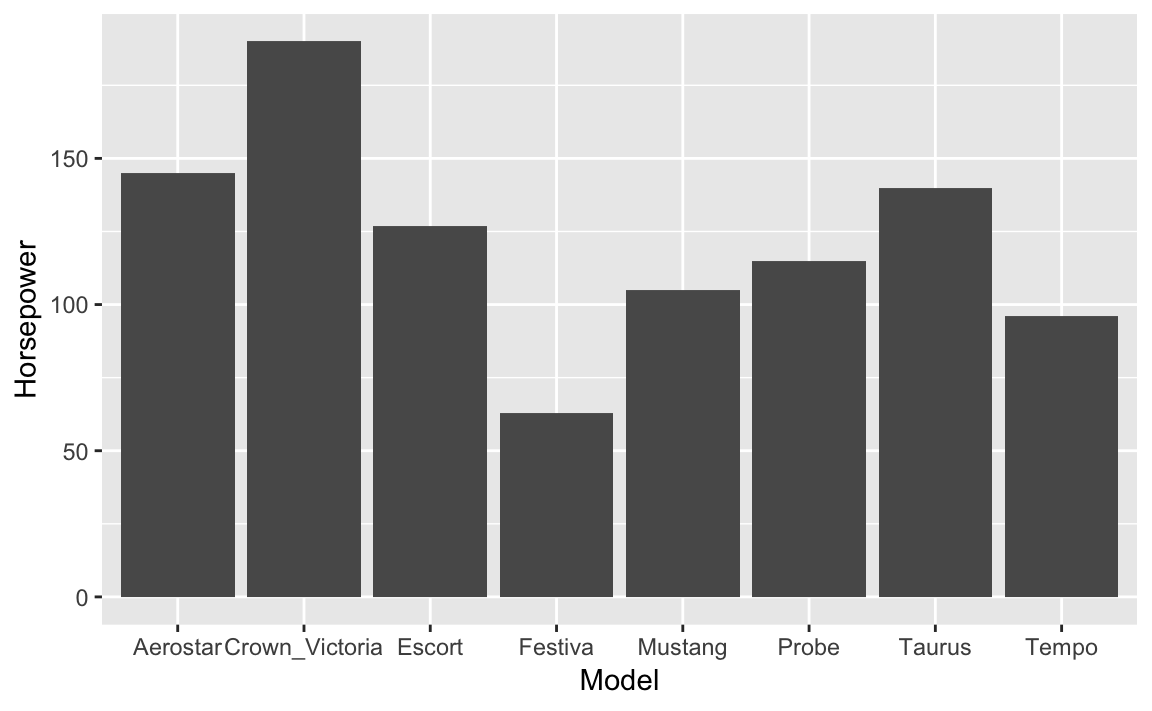
Figure 10-15. Ford Cars Bar Chart
The resulting graph in Figure 10-15 shows the resuting bar chart.
This example above uses stat = "identity" which assumes that the
heights of your bars are conveniently stored as a value in one field
with only one record per column. That is not always the case, however.
Often you have a vector of numeric data and a parallel factor or
character field that groups the data, and you want to produce a bar
chart of the group means or the group totals.
Let’s work up an example using the built-in airquality dataset which
contains daily temperature data for a single location for five months.
The data frame has a numeric Temp column and Month and Day
columns. If we want to plot the mean temp by month using ggplot we
don’t need to precompute the mean, instead we can have ggplot do that
in the plot command logic. To tell ggplot to calculate the mean we
pass stat = "summary", fun.y = "mean" to the geom_bar command. We
can also turn the month numbers into dates using the built in constant
month.abb which contains the abbreviations for the months.
ggplot(airquality,aes(month.abb[Month],Temp))+geom_bar(stat="summary",fun.y="mean")+labs(title="Mean Temp by Month",x="",y="Temp (deg. F)")
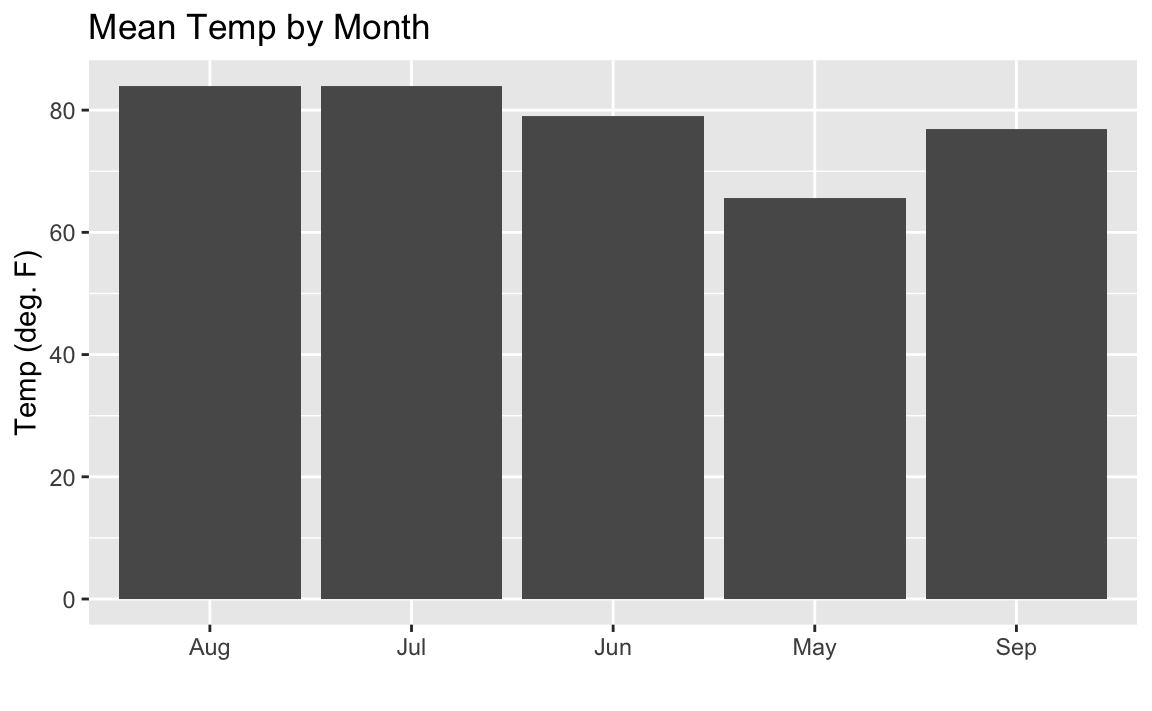
Figure 10-16. Bar Chart: Temp by Month
Figure 10-16 shows the resulting plot. But you might notice the sort order on the months is alphabetical, which is not how we typically like to see months sorted.
We can fix the sorting issue using a few functions from dplyr combined
with fct_inorder from the forcats Tidyverse package. To get the
months in the correct order we can sort the data frame by Month which
is the month number, then we can apply fct_inorder which will arrange
our factors in the order they appear in the data. You can see in Figure 10-17 that the bars are now sorted properly.
aq_data<-airquality%>%arrange(Month)%>%mutate(month_abb=fct_inorder(month.abb[Month]))ggplot(aq_data,aes(month_abb,Temp))+geom_bar(stat="summary",fun.y="mean")+labs(title="Mean Temp by Month",x="",y="Temp (deg. F)")
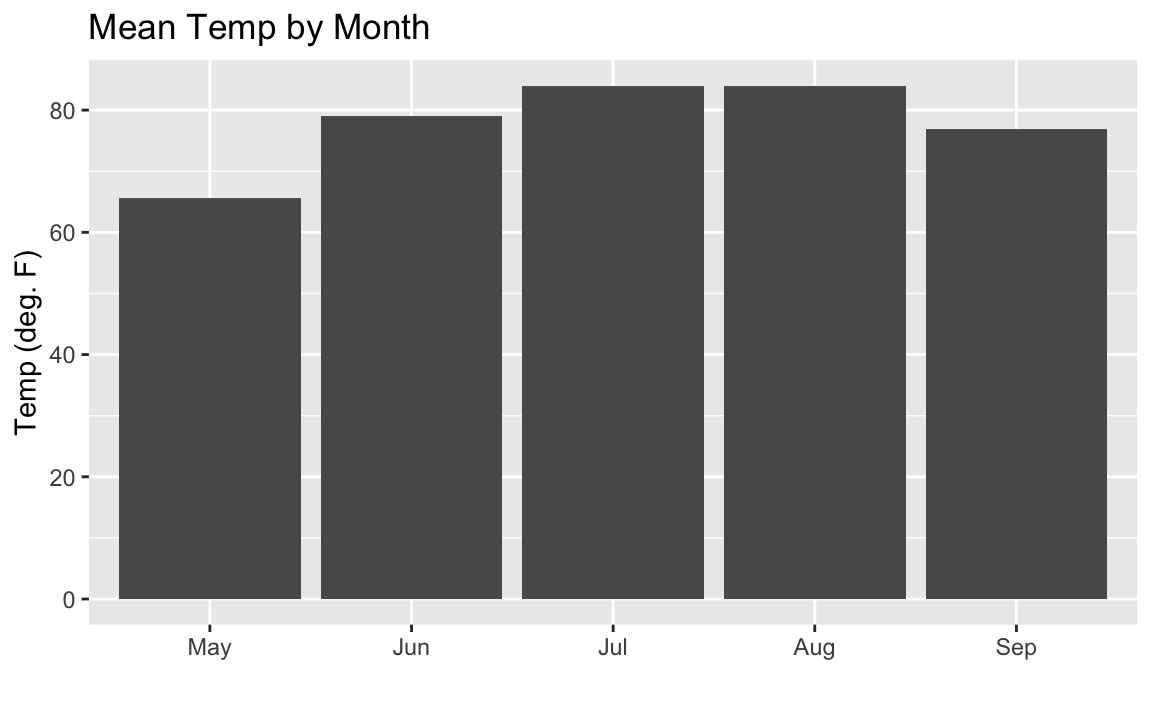
Figure 10-17. Bar Chart Properly Sorted
See Also
See “Adding Confidence Intervals to a Bar Chart” for adding confidence intervals and “Coloring a Bar Chart” for adding color
?geom_bar for help with bar charts in ggplot
barplot for Base R bar charts or the barchart function in the
lattice package.
Adding Confidence Intervals to a Bar Chart
Problem
You want to augment a bar chart with confidence intervals.
Solution
Suppose you have a data frame df with columns group which are group
names, stat which is a column of statistics, and lower and upper
which represent the corresponding limits for the confidence intervals.
We can display a bar chart of stat for each group and its confidence
intervals using the geom_bar combined with geom_errorbar.
ggplot(df,aes(group,stat))+geom_bar(stat="identity")+geom_errorbar(aes(ymin=lower,ymax=upper),width=.2)
. image::images/10_Graphics_files/figure-html/confbars-1.png[]
??? shows the resulting bar chart with confidence intervals.
Discussion
Most bar charts display point estimates, which are shown by the heights of the bars, but rarely do they include confidence intervals. Our inner statisticians dislike this intensely. The point estimate is only half of the story; the confidence interval gives the full story.
Fortunately, we can plot the error bars using ggplot. The hard part is
calculating the intervals. In the examples above our data had a simple
-15% and +20% interval. However, in “Creating a Bar Chart”, we calculated group means before plotting them. If we let
ggplot do the calculations for us we can use the build in mean_se
along with the stat_summary function to get the standard errors of the
mean measures.
Let’s use the airquality data we used previously. First we’ll do the
sorted factor procedure (from the prior recipe) to get the month names
in the desired order:
aq_data<-airquality%>%arrange(Month)%>%mutate(month_abb=fct_inorder(month.abb[Month]))
Now we can plot the bars along with the associated standard errors as in the following:
ggplot(aq_data,aes(month_abb,Temp))+geom_bar(stat="summary",fun.y="mean",fill="cornflowerblue")+stat_summary(fun.data=mean_se,geom="errorbar")+labs(title="Mean Temp by Month",x="",y="Temp (deg. F)")
Sometimes you’ll want to sort your columns in your bar chart in
descending order based on their height. This can be a little bit
confusing when using summary stats in ggplot but the secret is to use
mean in the reorder statement to sort the factor by the mean of the
temp. Note that the reference to mean in reorder is not quoted,
while the reference to mean in geom_bar is quoted:
ggplot(aq_data,aes(reorder(month_abb,-Temp,mean),Temp))+geom_bar(stat="summary",fun.y="mean",fill="tomato")+stat_summary(fun.data=mean_se,geom="errorbar")+labs(title="Mean Temp by Month",x="",y="Temp (deg. F)")
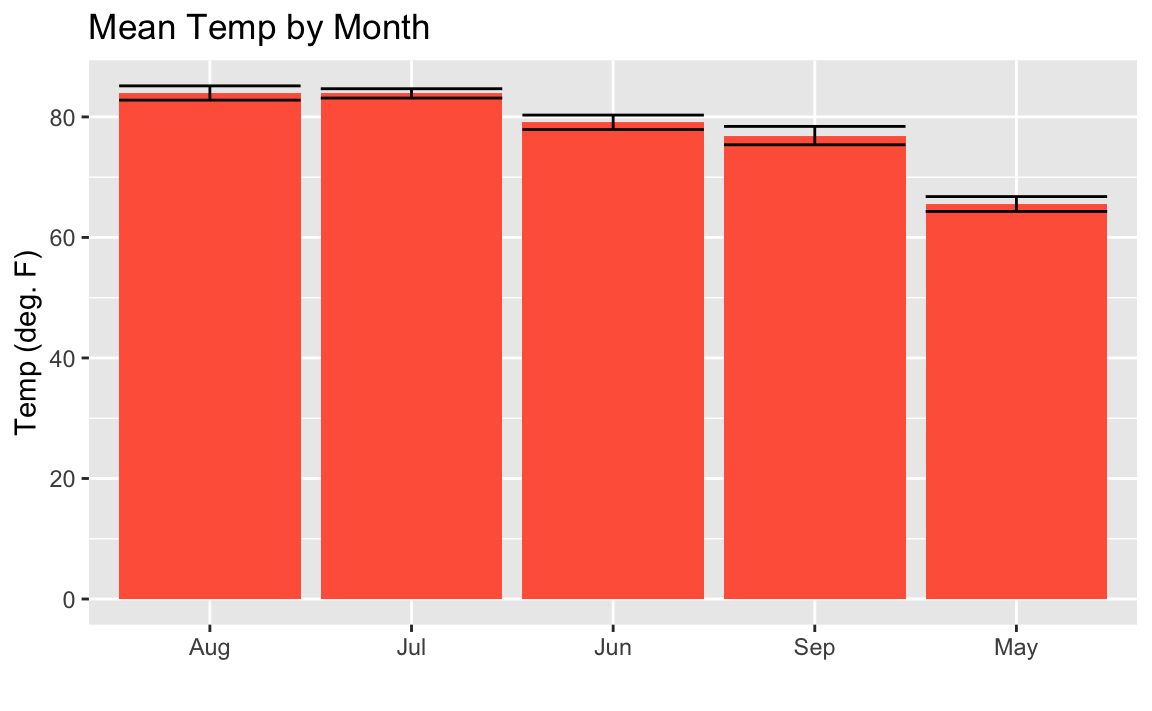
Figure 10-18. Mean Temp By Month Descending Order
You may look at this example and the result in Figure 10-18
and wonder, “Why didn’t they just use reorder(month_abb, Month) in the
first example instead of that sorting business with
forcats::fct_inorder to get the months in the right order?” Well, we
could have. But sorting using fct_inorder is a design pattern that
provides flexibility for more complicated things. Plus it’s quite easy
to read in a script. Using reorder inside the aes is a bit more
dense and hard to read later. But either approach is reasonable.
See Also
See “Forming a Confidence Interval for a Mean” for
more about t.test.
Coloring a Bar Chart
Problem
You want to color or shade the bars of a bar chart.
Solution
With gplot we add the fill= call to our aes and let ggplot pick
the colors for us:
ggplot(df,aes(x,y,fill=group))
Discussion
In ggplot we can use the fill parameter in aes to tell ggplot
what field to base the colors on. If we pass a numeric field to ggplot
we will get a continuous gradient of colors and if we pass a factor or
character field to fill we will get contrasting colors for each group.
Below we pass the character name of each month to the fill parameter:
aq_data<-airquality%>%arrange(Month)%>%mutate(month_abb=fct_inorder(month.abb[Month]))ggplot(data=aq_data,aes(month_abb,Temp,fill=month_abb))+geom_bar(stat="summary",fun.y="mean")+labs(title="Mean Temp by Month",x="",y="Temp (deg. F)")+scale_fill_brewer(palette="Paired")
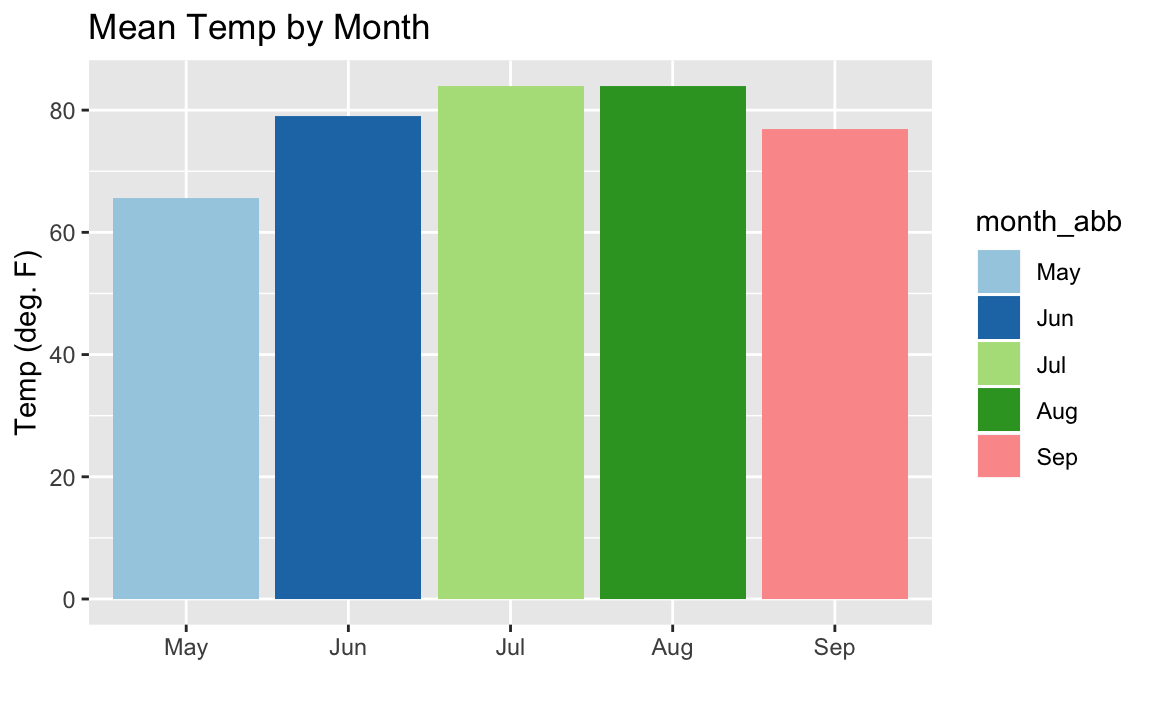
Figure 10-19. Colored Monthly Temp Bar Chart
The colors in the resulting Figure 10-19 are defined by
calling scale_fill_brewer(palette="Paired"). The "Paired" color
palette comes, along with many other color pallets, in the package
RColorBrewer.
If we wanted to change the color of each bar based on the temperature,
we can’t just set fill=Temp as might seem intuitive because ggplot
would not understand we want the mean temperature after the grouping by
month. So the way we get around this is to access a special field inside
of our graph called ..y.. which is the calculated value on the Y axis.
But we don’t want the legend labeled ..y.. so we add fill="Temp" to
our labs call in order to change the name of the legend. The result is
???
ggplot(airquality,aes(month.abb[Month],Temp,fill=..y..))+geom_bar(stat="summary",fun.y="mean")+labs(title="Mean Temp by Month",x="",y="Temp (deg. F)",fill="Temp")
images::images/10_Graphics_files/figure-latex/barsshaded-1.png[]
If we want to reverse the color scale, we can just add a negative - in
front of the field we are filling by. Like fill= -..y.., for example.
See Also
See “Creating a Bar Chart” for creating a bar chart.
Plotting a Line from x and y Points
Problem
You have paired observations in a data frame: (x1, y1), (x2, y2), …, (xn, yn). You want to plot a series of line segments that connect the data points.
Solution
With ggplot we can use geom_point to plot the points:.
ggplot(df,aes(x,y))+geom_point()
Since ggplot graphics are built up, element by element, we can have
both a point and a line in the same graphic very easily by having two
geoms:
ggplot(df,aes(x,y))+geom_point()+geom_line()
Discussion
To illustrate, let’s look at some example US economic data that comes
with ggplot2. This example data frame has a column called date which
we’ll plot on the x axis and a field unemploy which is the number of
unemployed people.
ggplot(economics,aes(date,unemploy))+geom_point()+geom_line()
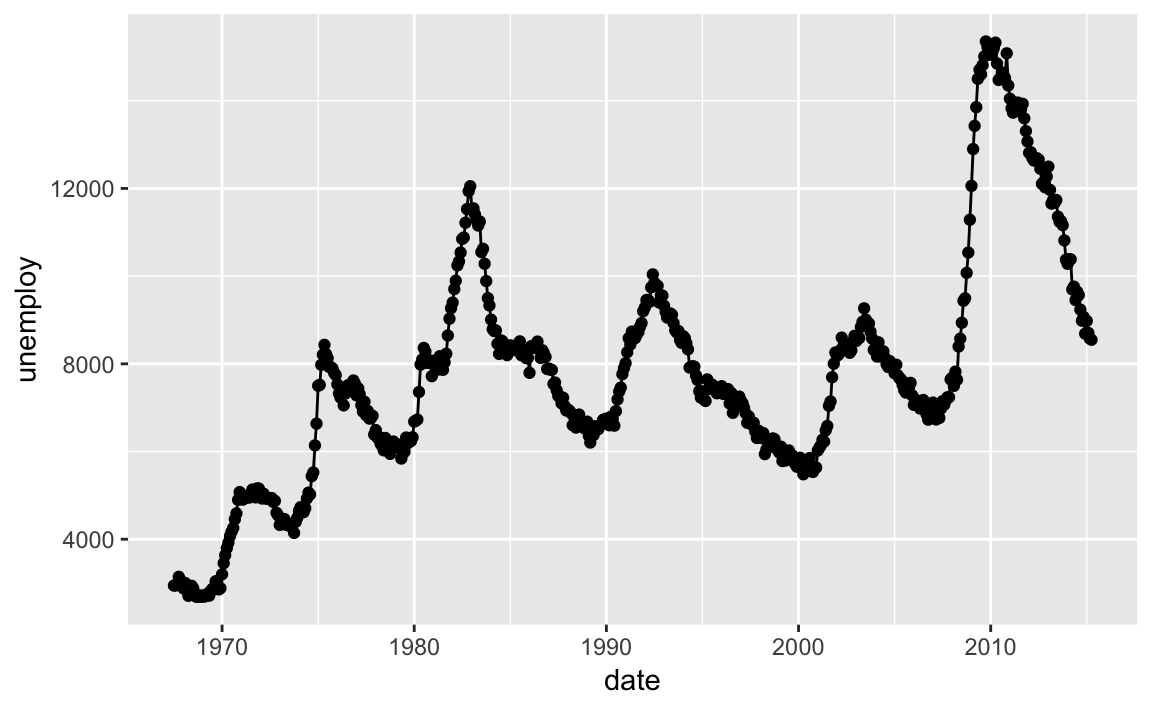
Figure 10-20. Line Chart Example
Figure 10-20 shows the resulting chart which contains both lines and points because we used both geoms.
See Also
Changing the Type, Width, or Color of a Line
Problem
You are plotting a line. You want to change the type, width, or color of the line.
Solution
ggplot uses the linetype parameter for controlling the appearance of
lines:
-
linetype="solid"orlinetype=1(default) -
linetype="dashed"orlinetype=2 -
linetype="dotted"orlinetype=3 -
linetype="dotdash"orlinetype=4 -
linetype="longdash"orlinetype=5 -
linetype="twodash"orlinetype=6 -
linetype="blank"orlinetype=0(inhibits drawing)
You can change the line characteristics by passing linetype, col
and/or size as parameters to the geom_line. So if we want to change
the linetype to dashed, red, and heavy we could pass the linetype,
col and size params to geom_line:
ggplot(df,aes(x,y))+geom_line(linetype=2,size=2,col="red")
Discussion
The example syntax above shows how to draw one line and specify its style, width, or color. A common scenario involves drawing multiple lines, each with its own style, width, or color.
Let’s set up some example data:
x<-1:10y1<-x**1.5y2<-x**2y3<-x**2.5df<-data.frame(x,y1,y2,y3)
In ggplot this can be a conundrum for many users. The challenge is
that ggplot works best with “long” data instead of “wide” data as was
mentioned in the introduction to this chapter. Our example data frame
has 4 columns of wide data:
head(df,3)#> x y1 y2 y3#> 1 1 1.00 1 1.00#> 2 2 2.83 4 5.66#> 3 3 5.20 9 15.59
We can make our wide data long by using the gather function from the
core tidyverse pacakge tidyr. In the example below, we use gather to
create a new column named bucket and put our column names in there
while keeping our x and y variables.
df_long<-gather(df,bucket,y,-x)head(df_long,3)#> x bucket y#> 1 1 y1 1.00#> 2 2 y1 2.83#> 3 3 y1 5.20tail(df_long,3)#> x bucket y#> 28 8 y3 181#> 29 9 y3 243#> 30 10 y3 316
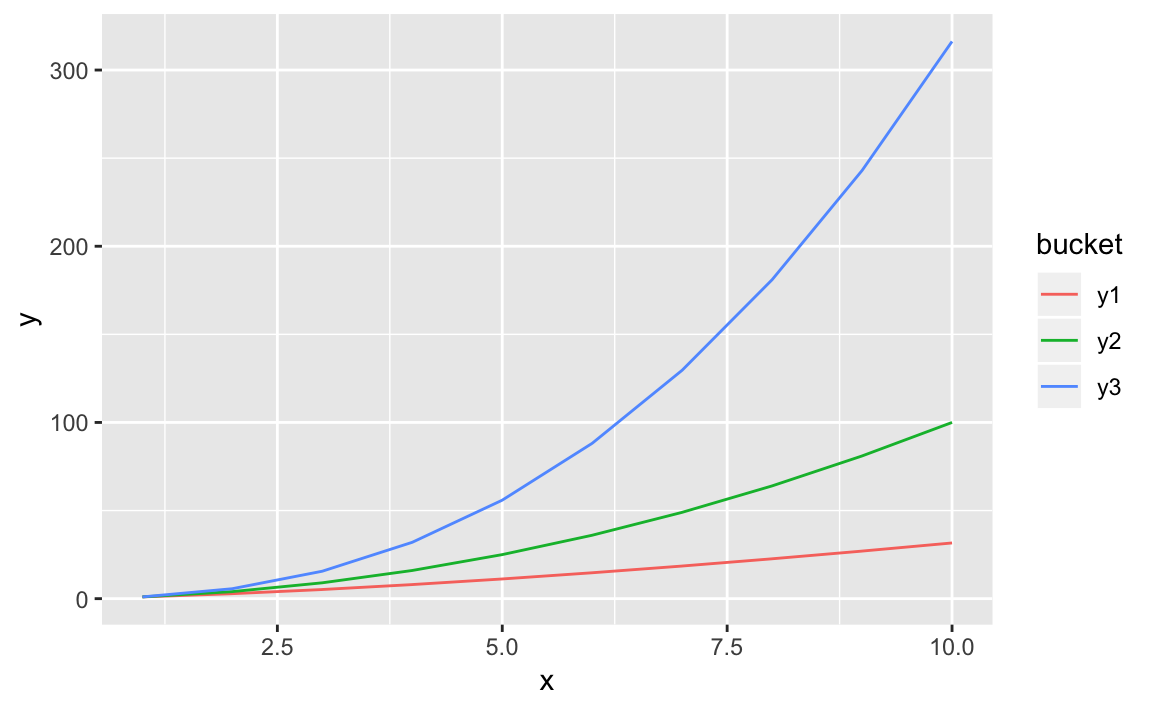
Now we can pass bucket to the col parameter and get multiple lines,
each a different color:
ggplot(df_long,aes(x,y,col=bucket))+geom_line()
It’s straight forward to vary the line weight by a variable by passing a
numerical variable to size:
ggplot(df,aes(x,y1,size=y2))+geom_line()+scale_size(name="Thickness based on y2")
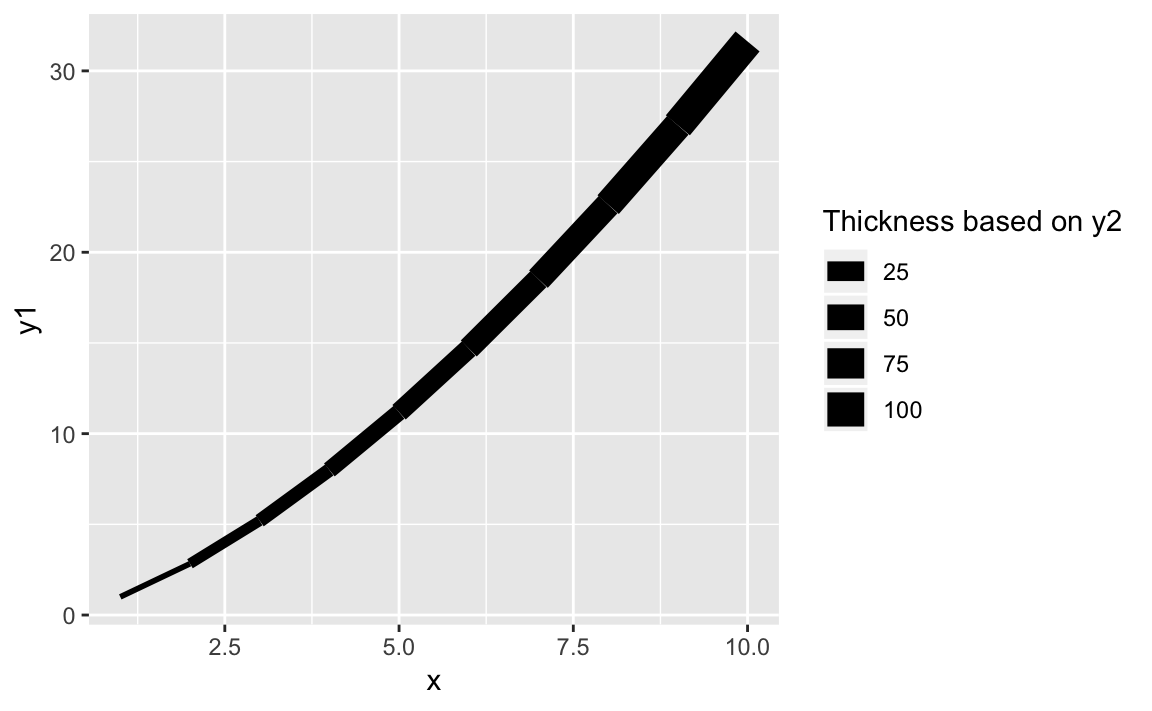
Figure 10-21. Thickness as a Function of x
The result of varying the thickness with x is shown in Figure 10-21.
See Also
See “Plotting a Line from x and y Points” for plotting a basic line.
Plotting Multiple Datasets
Problem
You want to show multiple datasets in one plot.
Solution
We could combine the data into one data frame before plotting using one
of the join functions from dplyr. However below we will create two
seperate data frames then add them each to a ggplot graph.
First let’s set up our example data frames, df1 and df2:
# example datan<-20x1<-1:ny1<-rnorm(n,0,.5)df1<-data.frame(x1,y1)x2<-(.5*n):((1.5*n)-1)y2<-rnorm(n,1,.5)df2<-data.frame(x2,y2)
Typically we would pass the data frame directly into the ggplot
function call. Since we want two geoms with two different data sources,
we will initiate a plot with ggplot() and then add in two calls to
geom_line each with its own data source.
ggplot()+geom_line(data=df1,aes(x=x1,y=y1),color="darkblue")+geom_line(data=df2,aes(x=x2,y=y2),linetype="dashed")
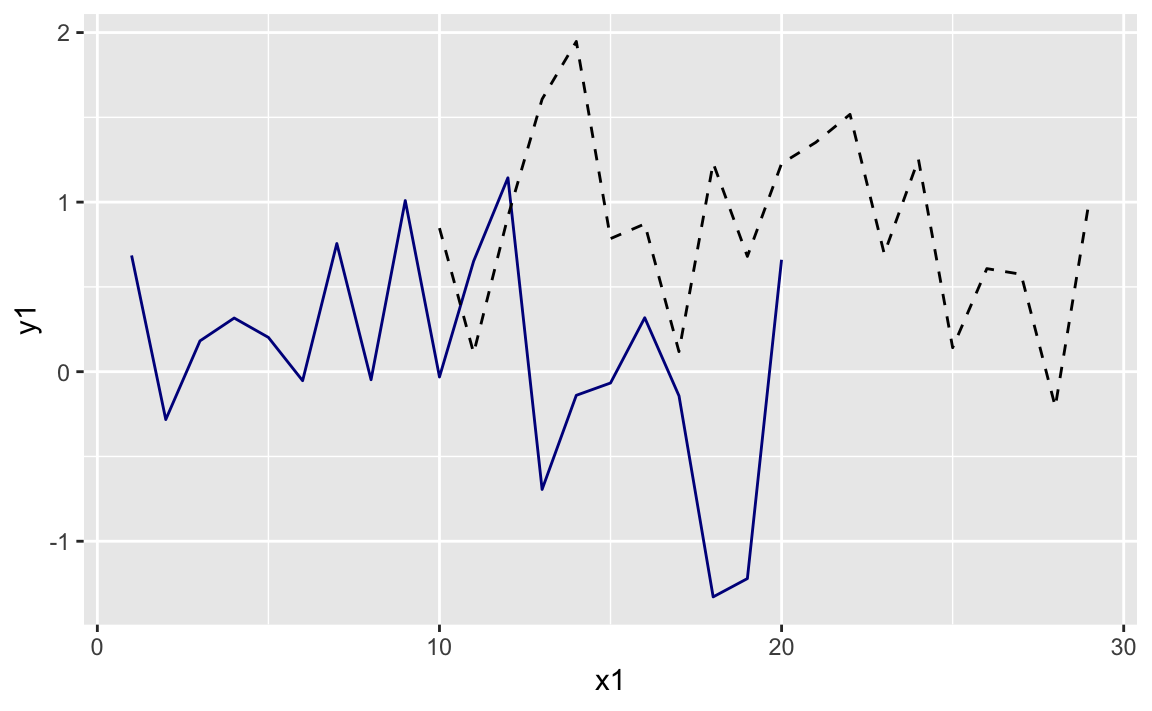
Figure 10-22. Two Lines One Plot
Discussion
ggplot allows us to make multiple calls to different geom_ functions
each with its own data source, if desired. Then ggplot will look at
all the data we are plotting and adjust the ranges to accomodate all the
data.
Even with good defaults, sometimes we want our plot range to show a
different range. We can do that by setting the xlim and ylim in our
ggplot.
ggplot()+geom_line(data=df1,aes(x=x1,y=y1),color="darkblue")+geom_line(data=df2,aes(x=x2,y=y2),linetype="dashed")+xlim(0,35)+ylim(-2,2)
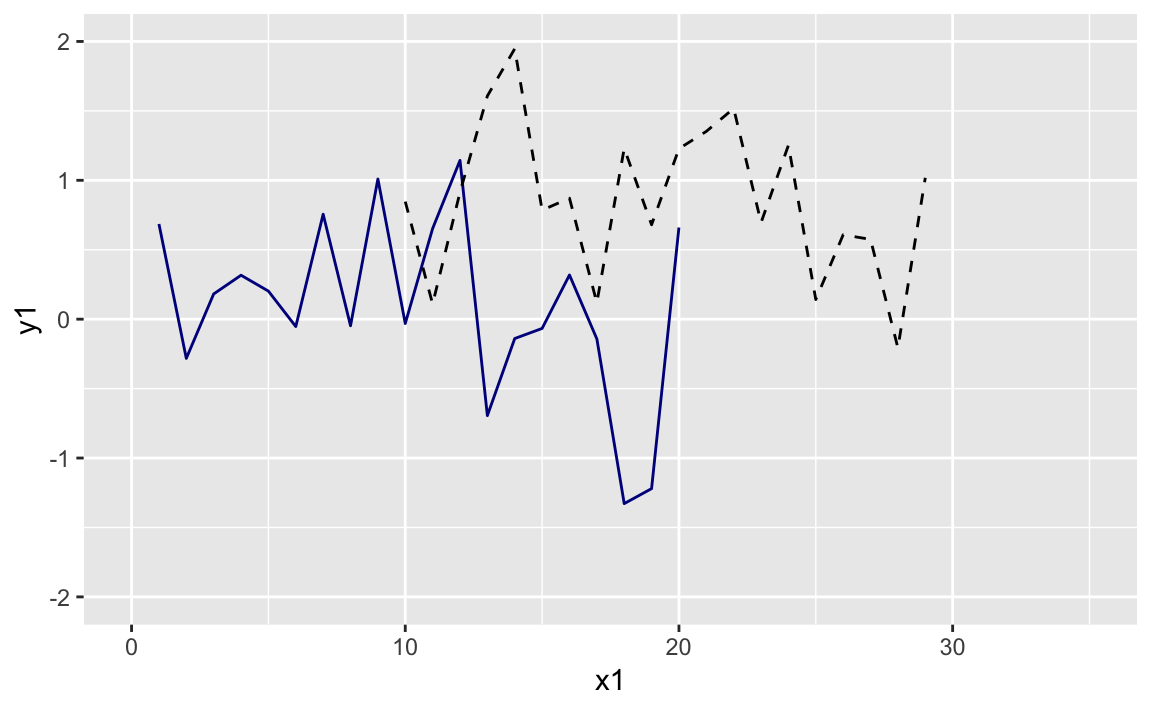
Figure 10-23. Two Lines Larger Limits
The graph with expanded limits is in Figure 10-23.
Adding Vertical or Horizontal Lines
Problem
You want to add a vertical or horizontal line to your plot, such as an axis through the origin or pointing out a threshold.
Solution
The ggplot functions geom_vline and geom_hline allow vertical and
horizontal lines, respectivly. The functions can also take color,
linetype, and size parameters to set the line style:
# using the data.frame df1 from the prior recipeggplot(df1)+aes(x=x1,y=y1)+geom_point()+geom_vline(xintercept=10,color="red",linetype="dashed",size=1.5)+geom_hline(yintercept=0,color="blue")
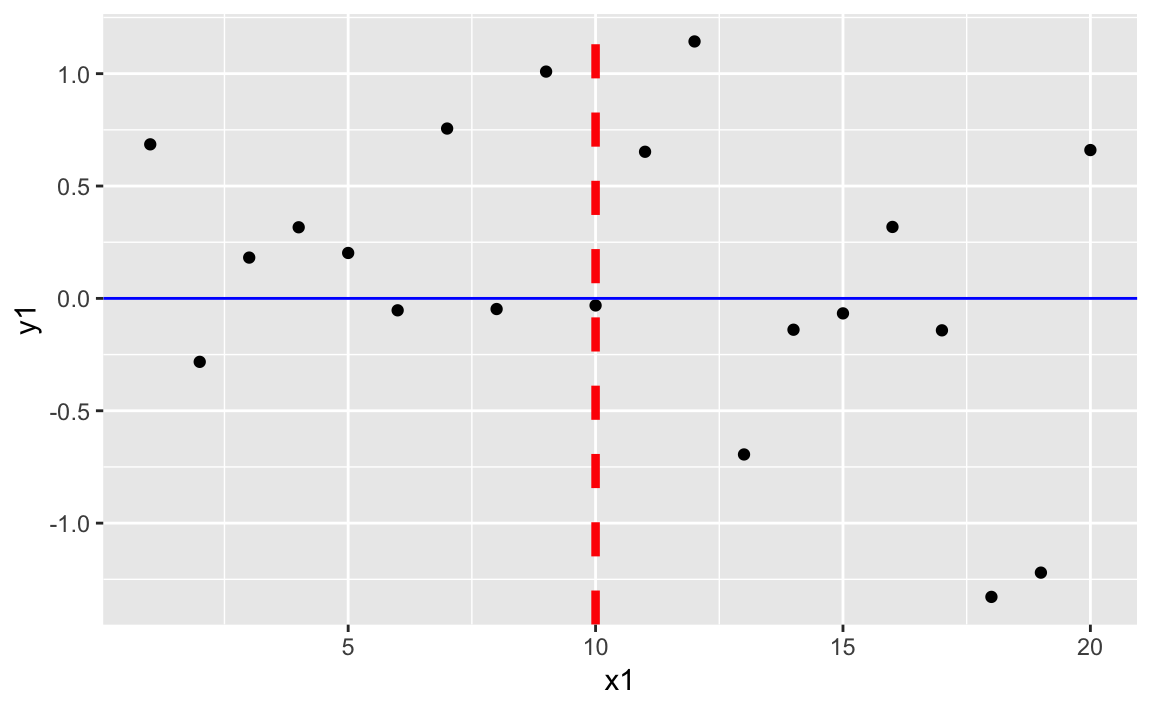
Figure 10-24. Vertical and Horizontal Lines
Figure 10-24 shows the resulting plot with added horizontal and vertical lines.
Discussion
A typical use of lines would be drawing regularly spaced lines. Suppose
we have a sample of points, samp. First, we plot them with a solid
line through the mean. Then we calculate and draw dotted lines at ±1 and
±2 standard deviations away from the mean. We can add the lines into our
plot with geom_hline:
samp<-rnorm(1000)samp_df<-data.frame(samp,x=1:length(samp))mean_line<-mean(samp_df$samp)sd_lines<-mean_line+c(-2,-1,+1,+2)*sd(samp_df$samp)ggplot(samp_df)+aes(x=x,y=samp)+geom_point()+geom_hline(yintercept=mean_line,color="darkblue")+geom_hline(yintercept=sd_lines,linetype="dotted")
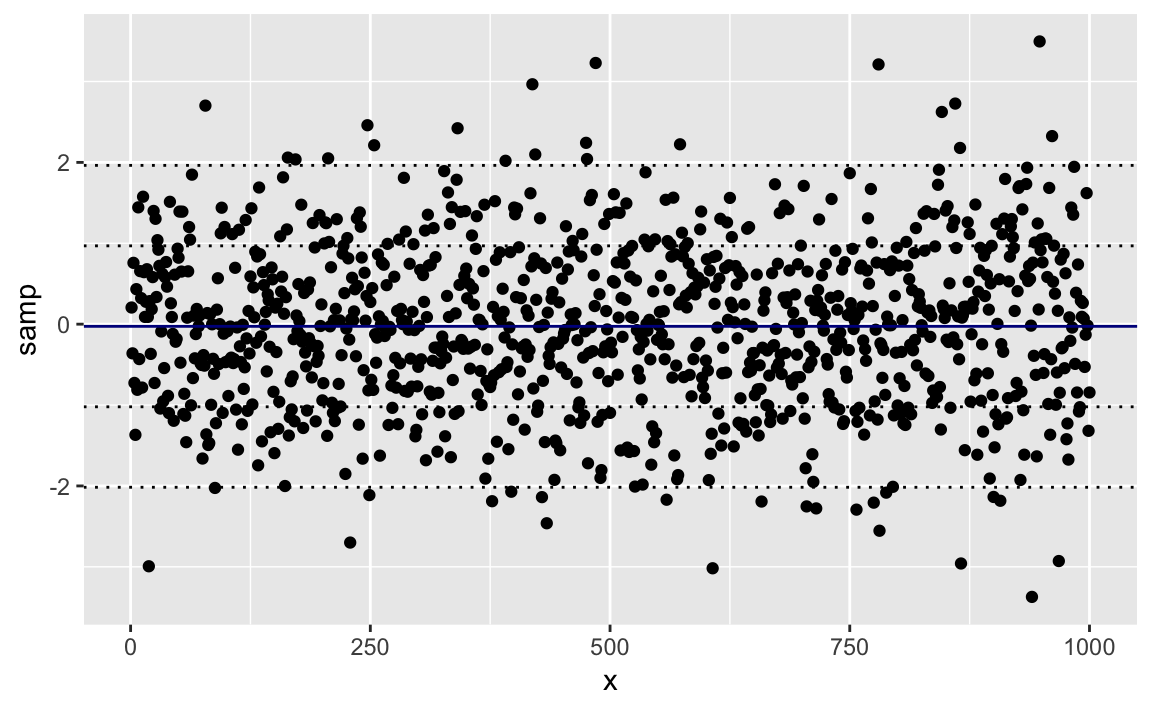
Figure 10-25. Mean and SD Bands in a Plot
Figure 10-25 shows the sampled data along with the mean and standard deviation lines.
See Also
See “Changing the Type, Width, or Color of a Line” for more about changing line types.
Creating a Box Plot
Problem
You want to create a box plot of your data.
Solution
Use geom_boxplot from ggplot to add a boxplot geom to a ggplot
graphic. Using the samp_df data frame from the prior recipe, we can
create a box plot of the values in the x column. The resulting graph
is in Figure 10-26.
ggplot(samp_df)+aes(y=samp)+geom_boxplot()
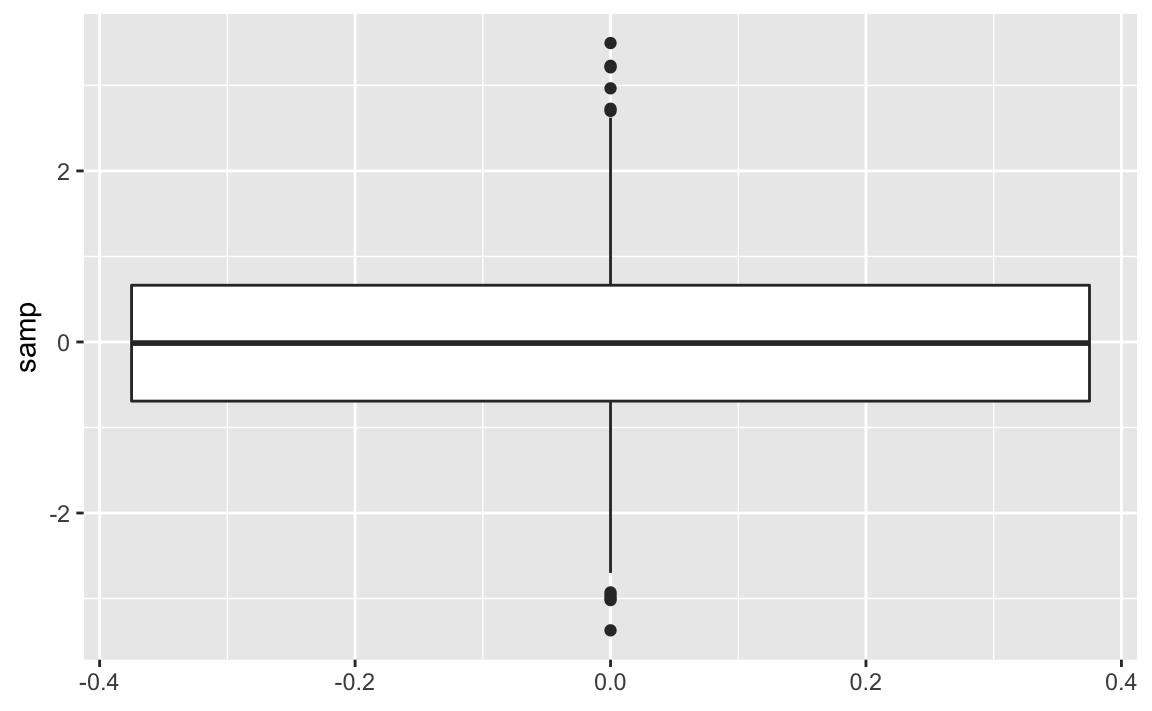
Figure 10-26. Single Boxplot
Discussion
A box plot provides a quick and easy visual summary of a dataset.
-
The thick line in the middle is the median.
-
The box surrounding the median identifies the first and third quartiles; the bottom of the box is Q1, and the top is Q3.
-
The “whiskers” above and below the box show the range of the data, excluding outliers.
-
The circles identify outliers. By default, an outlier is defined as any value that is farther than 1.5 × IQR away from the box. (IQR is the interquartile range, or Q3 − Q1.) In this example, there are a few outliers on the high side.
We can rotate the boxplot by flipping the coordinates. There are some situations where this makes a more appealing graphic. This is shown in Figure 10-27.
ggplot(samp_df)+aes(y=samp)+geom_boxplot()+coord_flip()
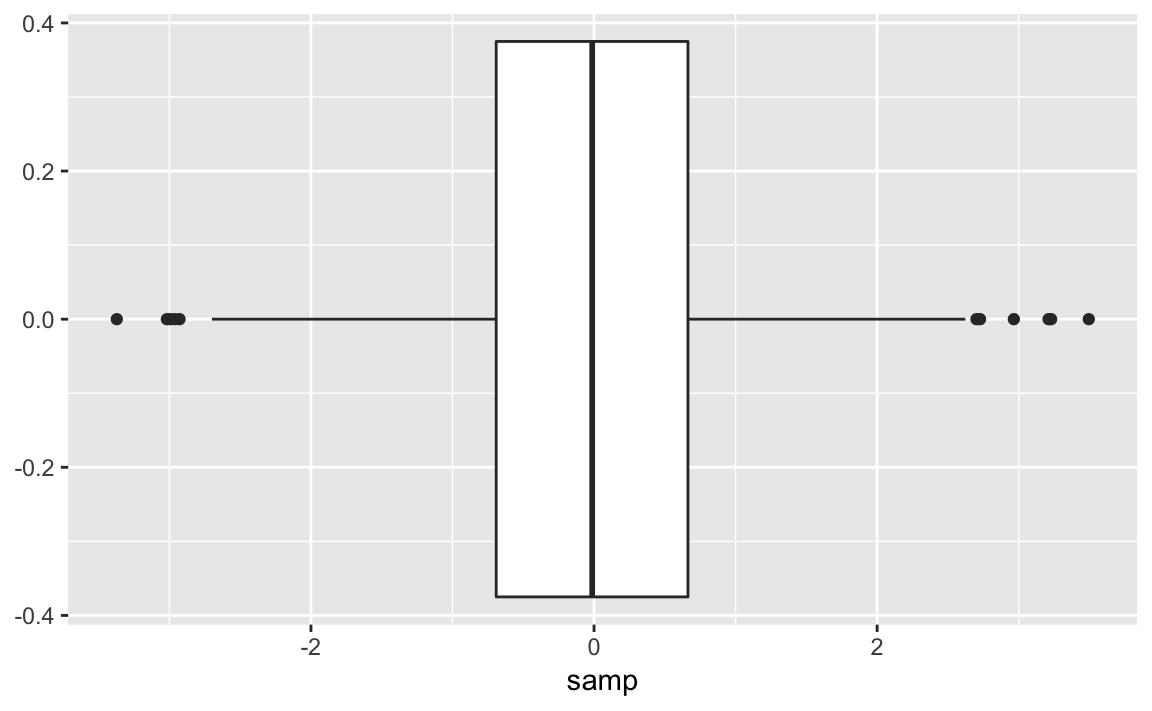
Figure 10-27. Single Boxplot
See Also
One box plot alone is pretty boring. See “Creating One Box Plot for Each Factor Level” for creating multiple box plots.
Creating One Box Plot for Each Factor Level
Problem
Your dataset contains a numeric variable and a factor (or other catagorical text). You want to create several box plots of the numeric variable broken out by levels.
Solution
With ggplot we pass the name of the categorical variable to the x
parameter in the aes call. The resulting boxplot will then be grouped
by the values in the categorical variable:
ggplot(df)+aes(x=factor,y=values)+geom_boxplot()
Discussion
This recipe is another great way to explore and illustrate the relationship between two variables. In this case, we want to know whether the numeric variable changes according to the level of a category.
The UScereal dataset from the MASS package contains many variables
regarding breakfast cereals. One variable is the amount of sugar per
portion and another is the shelf position (counting from the floor).
Cereal manufacturers can negotiate for shelf position, placing their
product for the best sales potential. We wonder: Where do they put the
high-sugar cereals? We can produce Figure 10-28 and
explore that question by creating one box plot per shelf:
data(UScereal,package="MASS")ggplot(UScereal)+aes(x=as.factor(shelf),y=sugars)+geom_boxplot()+labs(title="Sugar Content by Shelf",x="Shelf",y="Sugar (grams per portion)")
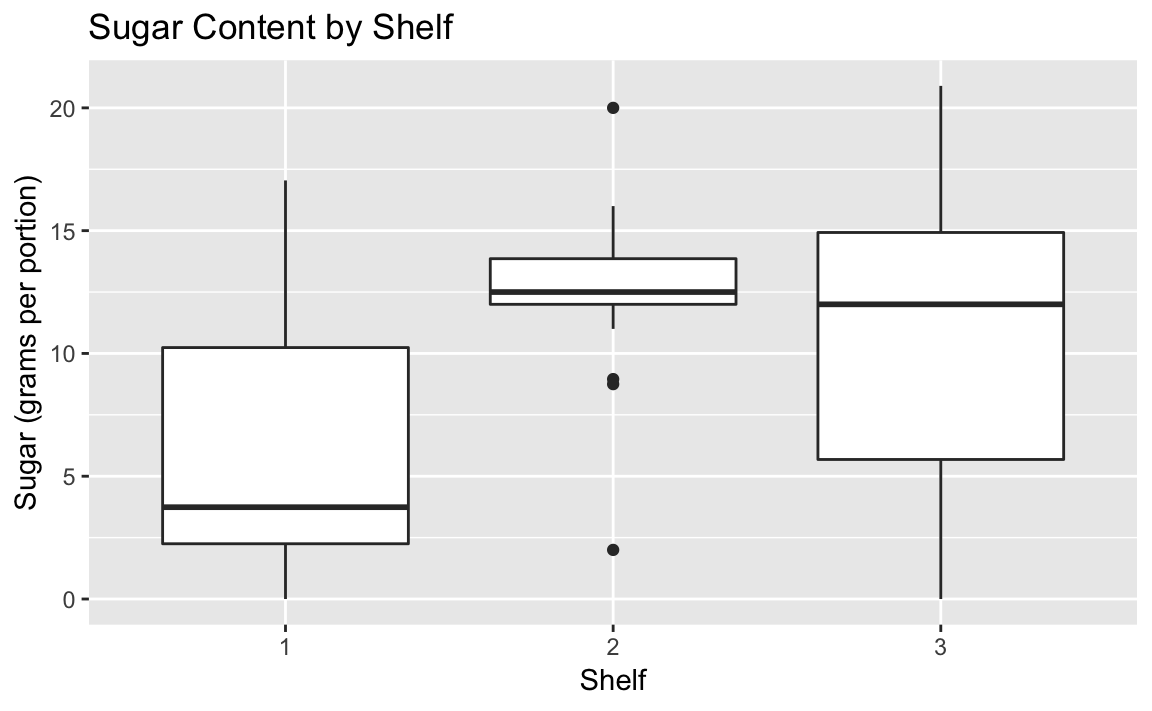
Figure 10-28. Boxplot by Shelf Number
The box plots suggest that shelf #2 has the most high-sugar cereals. Could it be that this shelf is at eye level for young children who can influence their parent’s choice of cereals?
Note that in the aes call we had to tell ggplot to treat the shelf
number as a factor. Otherwise ggplot would not react to the shelf as a
grouping and only print a single boxplot.
See Also
See “Creating a Box Plot” for creating a basic box plot.
Creating a Histogram
Problem
You want to create a histogram of your data.
Solution
Use geom_histogram, and set x to a vector of numeric values.
Discussion
Figure 10-29 is a histogram of the MPG.city column taken
from the Cars93 dataset:
data(Cars93,package="MASS")ggplot(Cars93)+geom_histogram(aes(x=MPG.city))#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
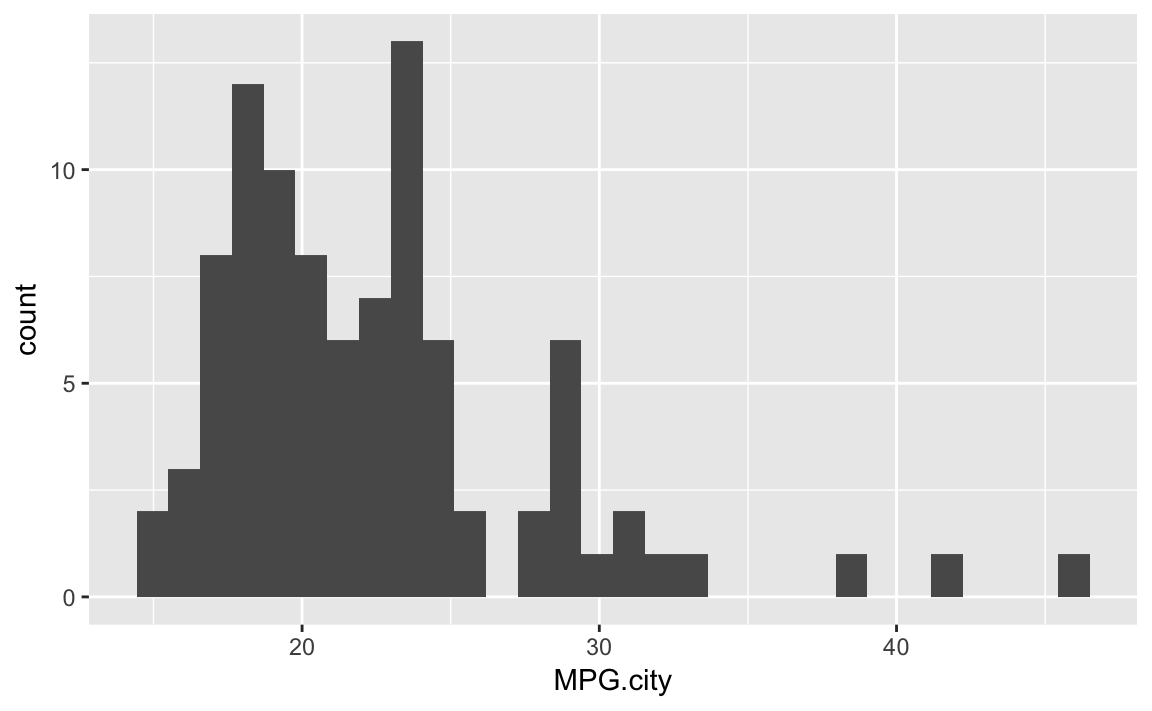
Figure 10-29. Histogram of Counts by MPG
The geom_histogram function must decide how many cells (bins) to
create for binning the data. In this example, the default algorithm
chose 30 bins. If we wanted fewer bins, we wo would include the bins
parameter to tell geom_histogram how many bins we want:
ggplot(Cars93)+geom_histogram(aes(x=MPG.city),bins=13)
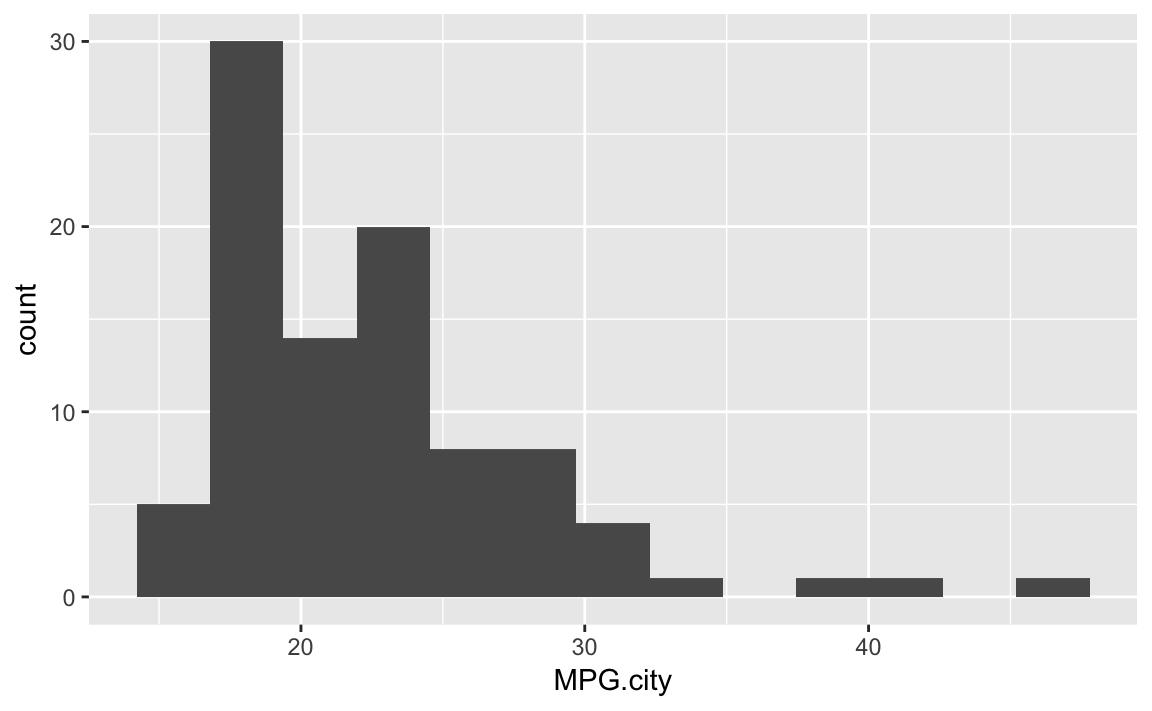
Figure 10-30. Histogram of Counts by MPG with Fewer Bins
Figure 10-30 shows the histogram with 13 bins.
See Also
The Base R function hist provides much of the same functionality as
does the histogram function of the lattice package.
Adding a Density Estimate to a Histogram
Problem
You have a histogram of your data sample, and you want to add a curve to illustrate the apparent density.
Solution
Use the geom_density function to approximate the sample density as
shown in Figure 10-31:
ggplot(Cars93)+aes(x=MPG.city)+geom_histogram(aes(y=..density..),bins=21)+geom_density()
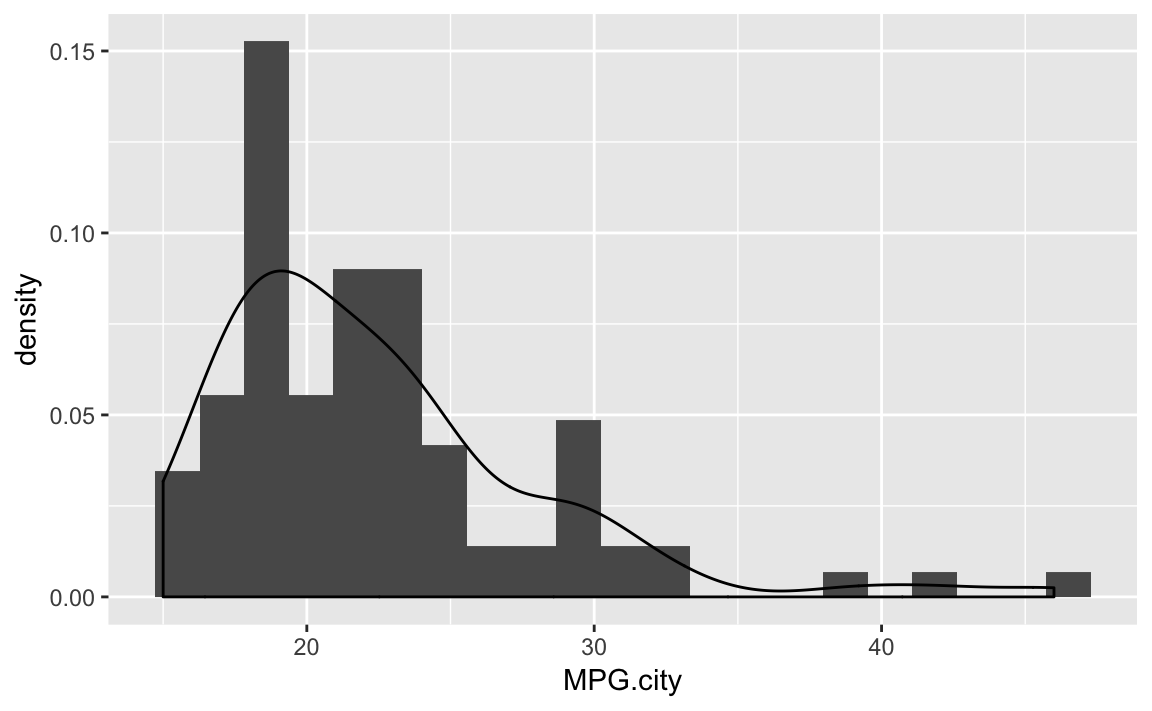
Figure 10-31. Histogram with Density Plot
Discussion
A histogram suggests the density function of your data, but it is rough. A smoother estimate could help you better visualize the underlying distribution. A Kernel Density Estimation (KDE) is a smoother representation of univariate data.
In ggplot we tell the geom_histogram function to use the density
function by passing it aes(y = ..density..).
The following example takes a sample from a gamma distribution and then plots the histogram and the estimated density as shown in Figure 10-32.
samp<-rgamma(500,2,2)ggplot()+aes(x=samp)+geom_histogram(aes(y=..density..),bins=10)+geom_density()
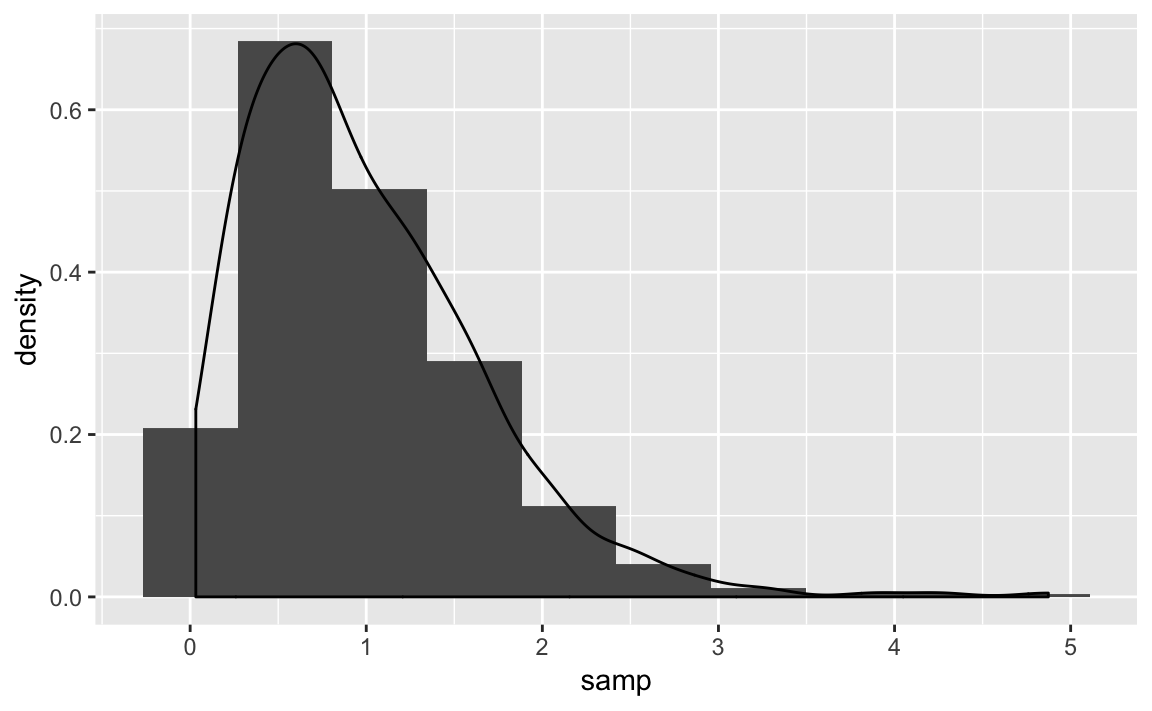
Figure 10-32. Histogram and Density: Gamma Distribution
See Also
The density function approximates the shape of the density nonparametrically. If you know the actual underlying distribution, use instead “Plotting a Density Function” to plot the density function.
Creating a Normal Quantile-Quantile (Q-Q) Plot
Problem
You want to create a quantile-quantile (Q-Q) plot of your data, typically because you want to know how the data differs from a normal distribution.
Solution
With ggplot we can use the stat_qq and stat_qq_line functions to
create a Q-Q plot that shows both the observed points as well as the Q-Q
Line. Figure 10-33 shows the resulting plot.
df<-data.frame(x=rnorm(100))ggplot(df,aes(sample=x))+stat_qq()+stat_qq_line()
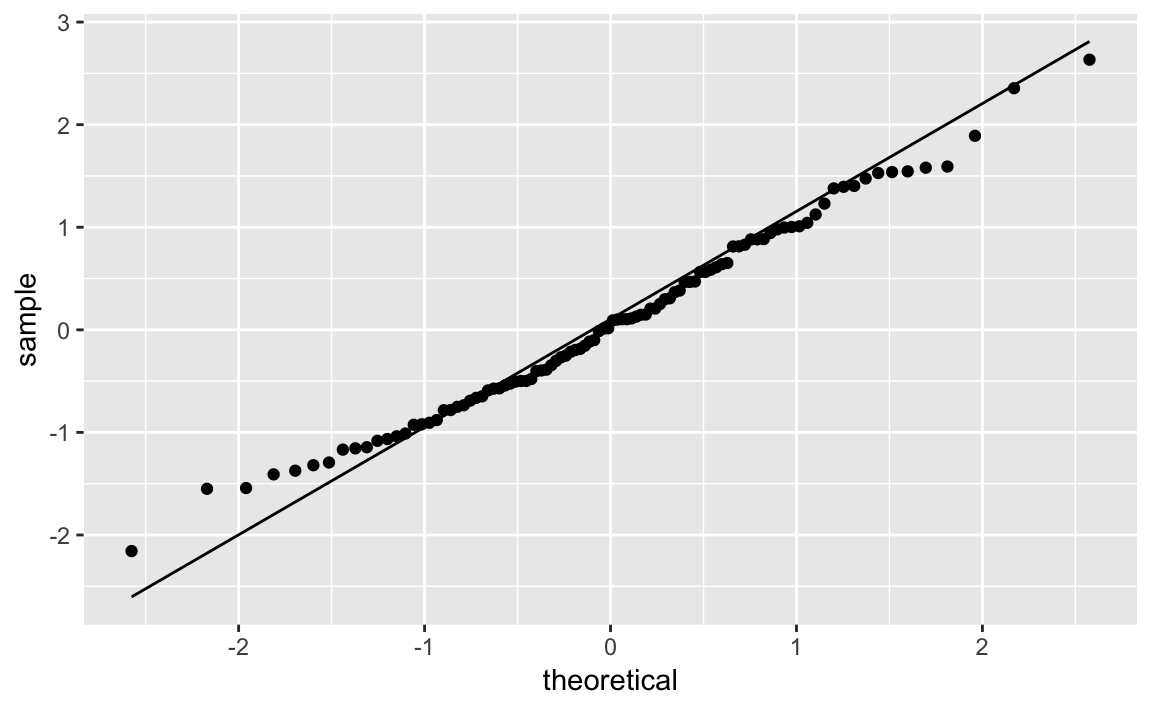
Figure 10-33. Q-Q Plot
Discussion
Sometimes it’s important to know if your data is normally distributed. A quantile-quantile (Q-Q) plot is a good first check.
The Cars93 dataset contains a Price column. Is it normally
distributed? This code snippet creates a Q-Q plot of Price shown in
Figure 10-34:
ggplot(Cars93,aes(sample=Price))+stat_qq()+stat_qq_line()
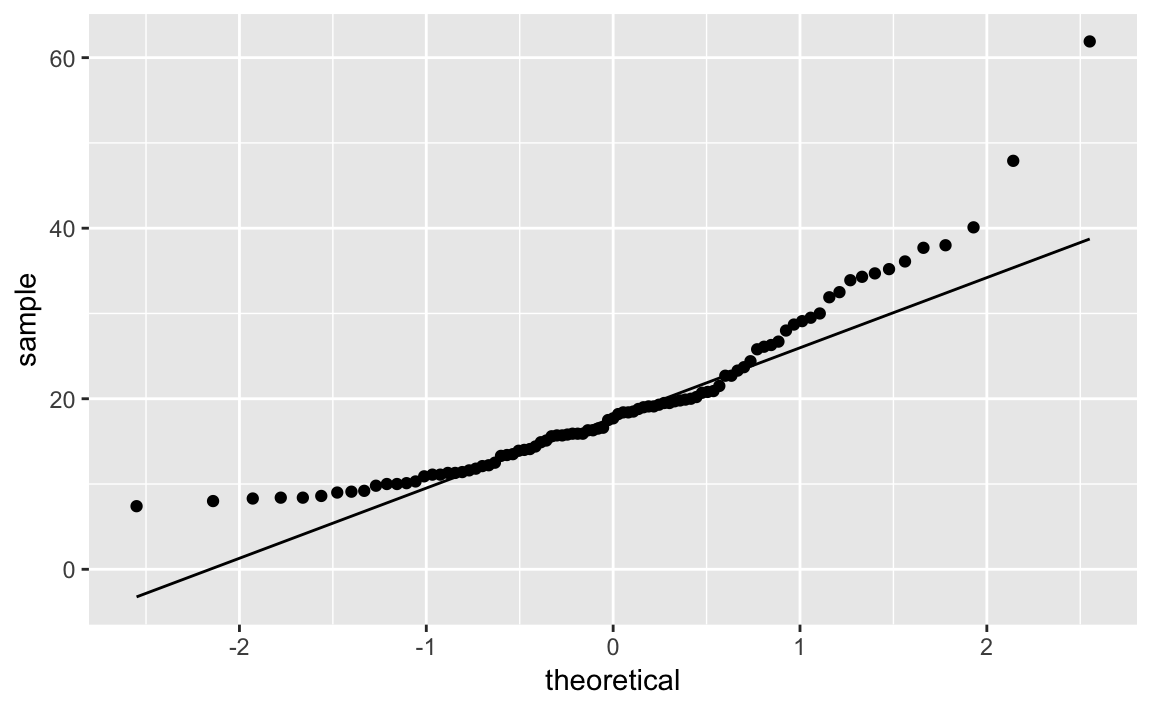
Figure 10-34. Q-Q Plot of Car Prices
If the data had a perfect normal distribution, then the points would fall exactly on the diagonal line. Many points are close, especially in the middle section, but the points in the tails are pretty far off. Too many points are above the line, indicating a general skew to the left.
The leftward skew might be cured by a logarithmic transformation. We can plot log(Price), which yields Figure 10-35:
ggplot(Cars93,aes(sample=log(Price)))+stat_qq()+stat_qq_line()
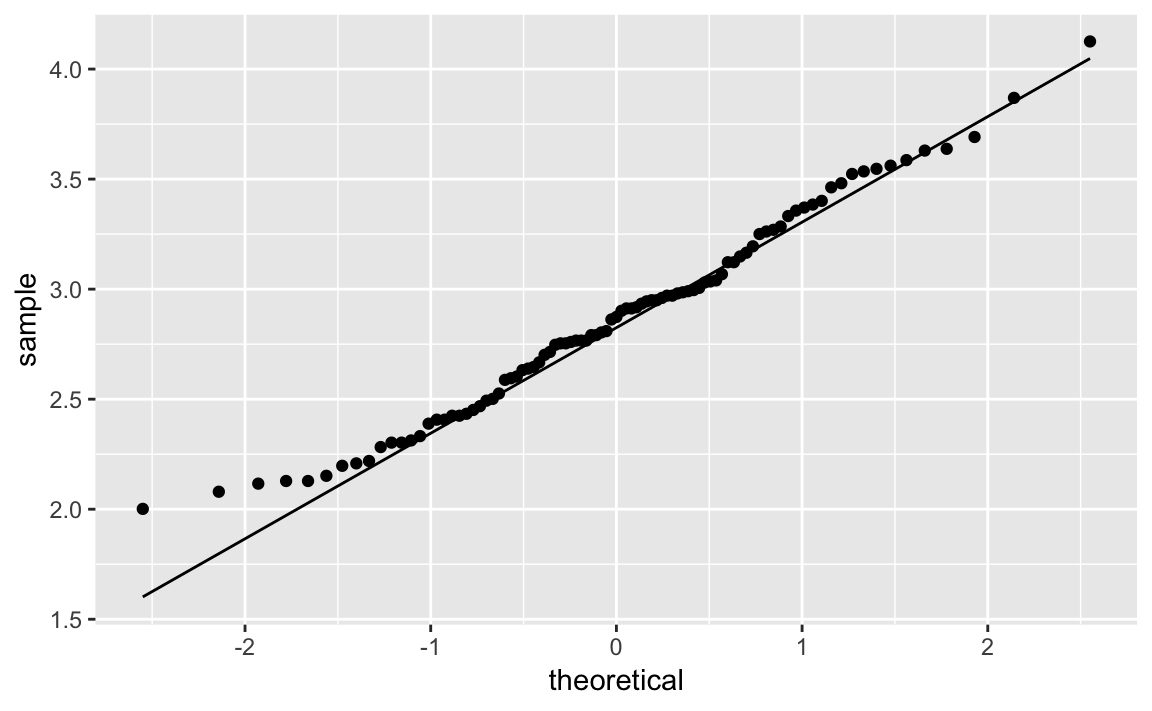
Figure 10-35. Q-Q Plot of Log Car Prices
Notice that the points in the new plot are much better behaved, staying close to the line except in the extreme left tail. It appears that log(Price) is approximately Normal.
See Also
See “Creating Other Quantile-Quantile Plots” for creating Q-Q plots for other distributions. See Recipe X-X for an application of Normal Q-Q plots to diagnosing linear regression.
Creating Other Quantile-Quantile Plots
Problem
You want to view a quantile-quantile plot for your data, but the data is not normally distributed.
Solution
For this recipe, you must have some idea of the underlying distribution, of course. The solution is built from the following steps:
-
Use the
ppointsfunction to generate a sequence of points between 0 and 1. -
Transform those points into quantiles, using the quantile function for the assumed distribution.
-
Sort your sample data.
-
Plot the sorted data against the computed quantiles.
-
Use
ablineto plot the diagonal line.
This can all be done in two lines of R code. Here is an example that
assumes your data, y, has a Student’s t distribution with 5 degrees
of freedom. Recall that the quantile function for Student’s t is qt
and that its second argument is the degrees of freedom. To create draws
from
First let’s make some example data:
df_t<-data.frame(y=rt(100,5))
In order to plot the Q-Q plot we need to estimate the parameters of the
distribution we’re wanting to plot. Since this is a Student’s t
distribution, we only need to estimate one parameter, the degrees of
freedom. Of course we know the actual degrees of freedom is 5, but in
most situations we’ll need to calcuate value. So we’ll use the
MASS::fitdistr function to estimate the degrees of freedom:
est_df<-as.list(MASS::fitdistr(df_t$y,"t")$estimate)[["df"]]#> Warning in log(s): NaNs produced#> Warning in log(s): NaNs produced#> Warning in log(s): NaNs producedest_df#> [1] 19.5
As expected, that’s pretty close to what was used to generate the simulated data. So let’s pass the estimaged degrees of freedom to the Q-Q functions and create Figure 10-36:
ggplot(df_t)+aes(sample=y)+geom_qq(distribution=qt,dparams=est_df)+stat_qq_line(distribution=qt,dparams=est_df)
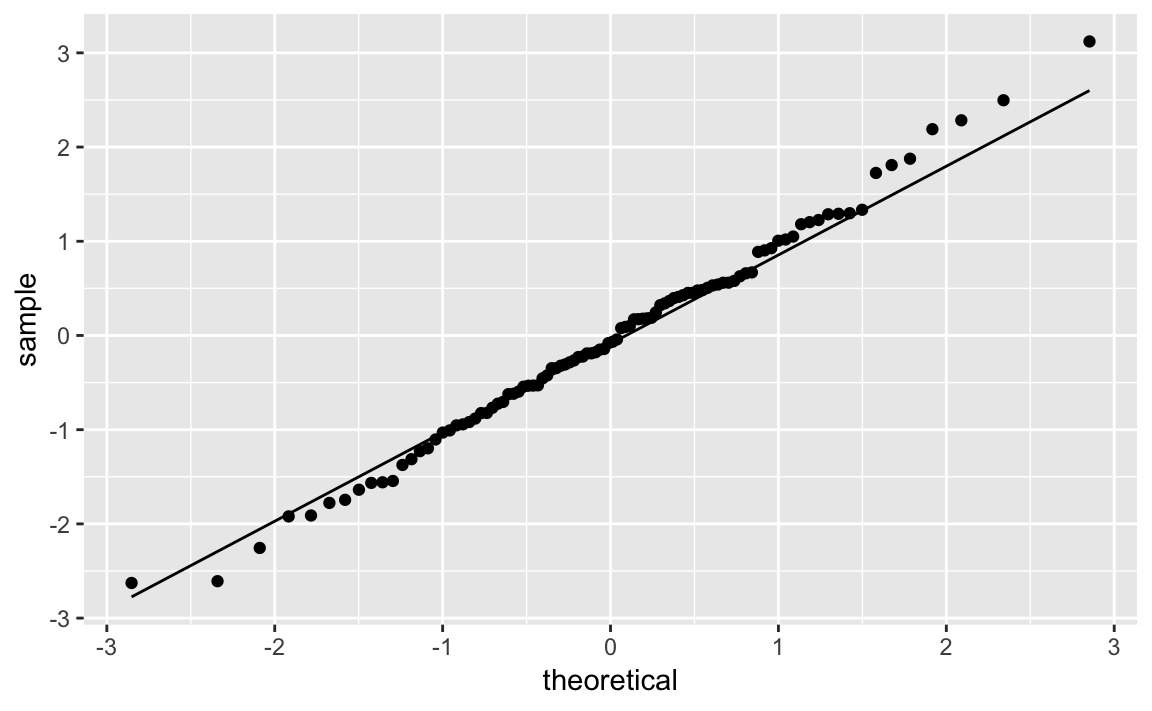
Figure 10-36. Student’s t Distribution Q-Q Plot
Discussion
The solution looks complicated, but the gist of it is picking a
distribution, fitting the parameters, and then passing those parameters
to the Q-Q functions in ggplot.
We can illustrate this recipe by taking a random sample from an exponential distribution with a mean of 10 (or, equivalently, a rate of 1/10):
rate<-1/10n<-1000df_exp<-data.frame(y=rexp(n,rate=rate))
est_exp<-as.list(MASS::fitdistr(df_exp$y,"exponential")$estimate)[["rate"]]est_exp#> [1] 0.101
Notice that for an exponential distribution the parameter we estimate is
called rate as opposed to df which was the parameter in the t
distribution.
ggplot(df_exp)+aes(sample=y)+geom_qq(distribution=qexp,dparams=est_exp)+stat_qq_line(distribution=qexp,dparams=est_exp)
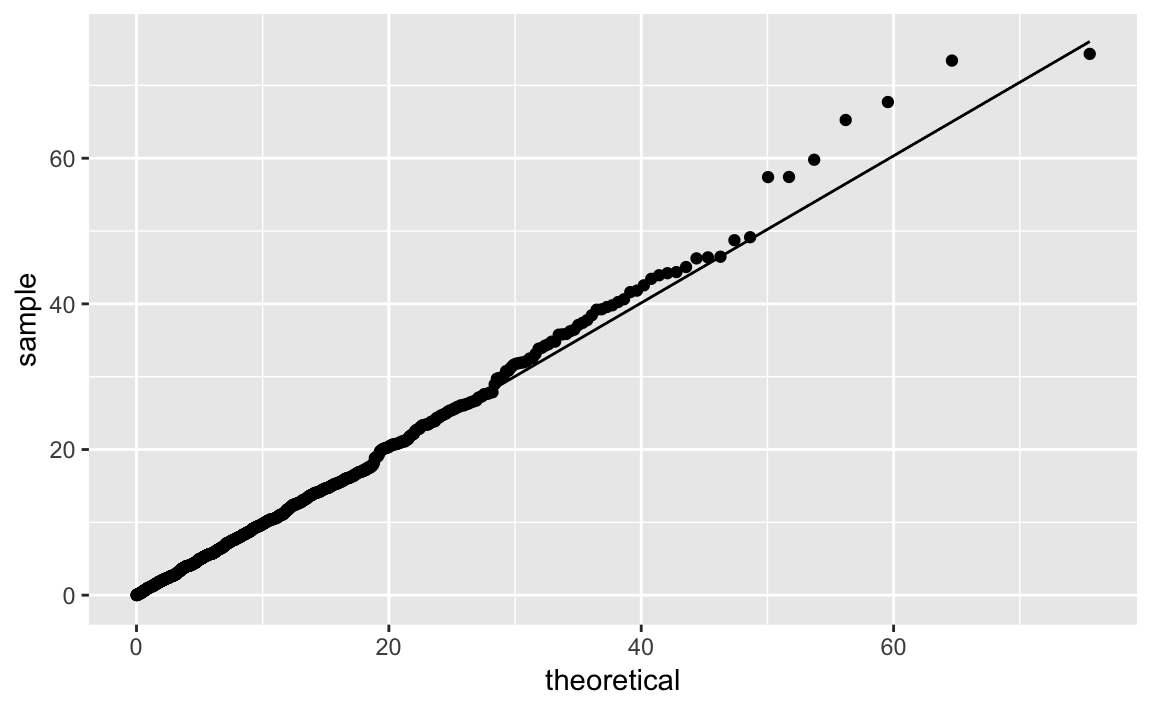
Figure 10-37. Exponential Distribution Q-Q Plot
The quantile function for the exponential distribution is qexp, which
takes the rate argument. Figure 10-37 shows the resulting
Q-Q plot using a theoretical exponential distribution.
Plotting a Variable in Multiple Colors
Problem
You want to plot your data in multiple colors, typically to make the plot more informative, readable, or interesting.
Solution
We can pass a color to a geom_ function in order to produced colored
output:
df<-data.frame(x=rnorm(200),y=rnorm(200))ggplot(df)+aes(x=x,y=y)+geom_point(color="blue")
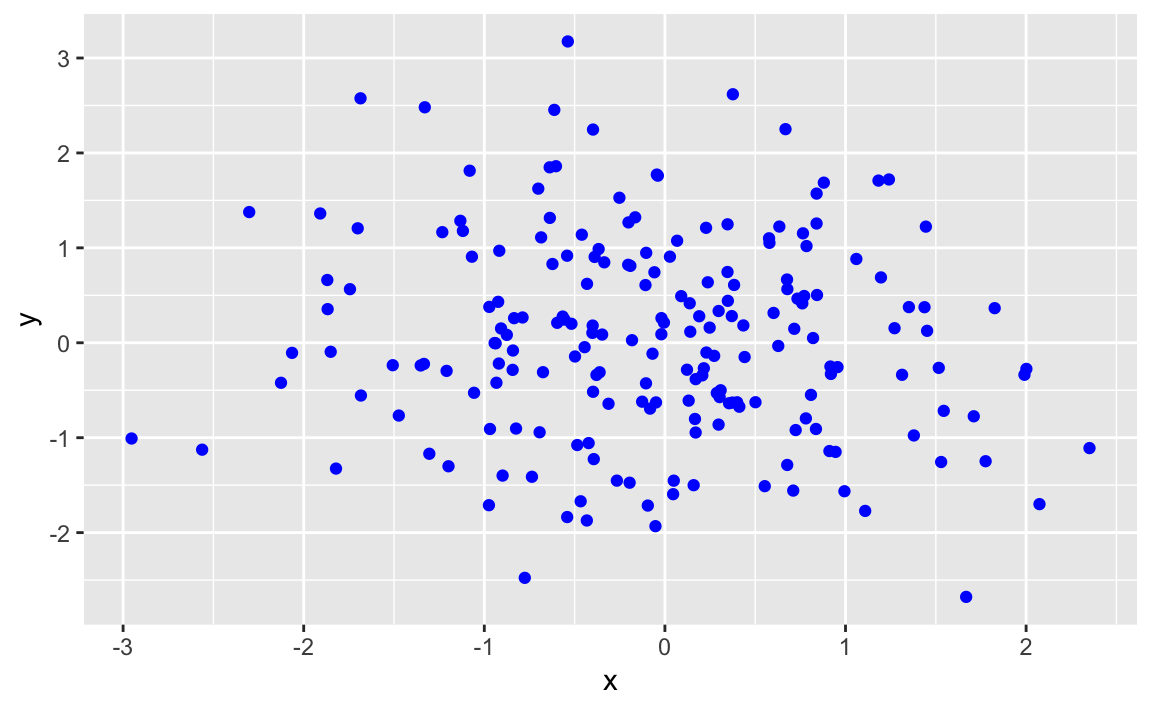
Figure 10-38. Point Data in Color
The value of color can be:
-
One color, in which case all data points are that color.
-
A vector of colors, the same length as
x, in which case each value ofxis colored with its corresponding color. -
A short vector, in which case the vector of colors is recycled.
Discussion
The default color in ggplot is black. While it’s not very exciting,
black is high contrast and easy for most anyone to see.
However, it is much more useful (and interesting) to vary the color in a way that illuminates the data. Let’s illustrate this by plotting a graphic two ways, once in black and white and once with simple shading.
This produces the basic black-and-white graphic in Figure 10-39:
df<-data.frame(x=1:100,y=rnorm(100))ggplot(df)+aes(x,y)+geom_point()
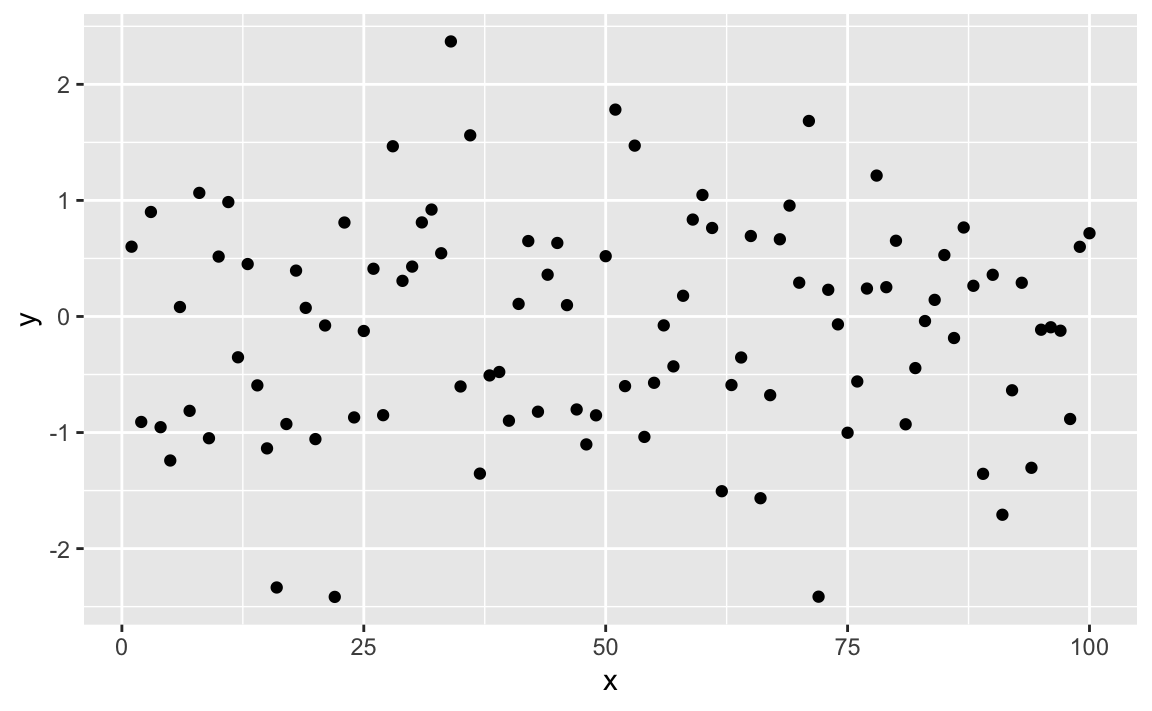
Figure 10-39. Simple Point Plot
Now we can make it more interesting by creating a vector of "gray" and
"black" values, according to the sign of x and then plotting x
using those colors as shown in Figure 10-40:
shade<-if_else(df$y>=0,"black","gray")ggplot(df)+aes(x,y)+geom_point(color=shade)
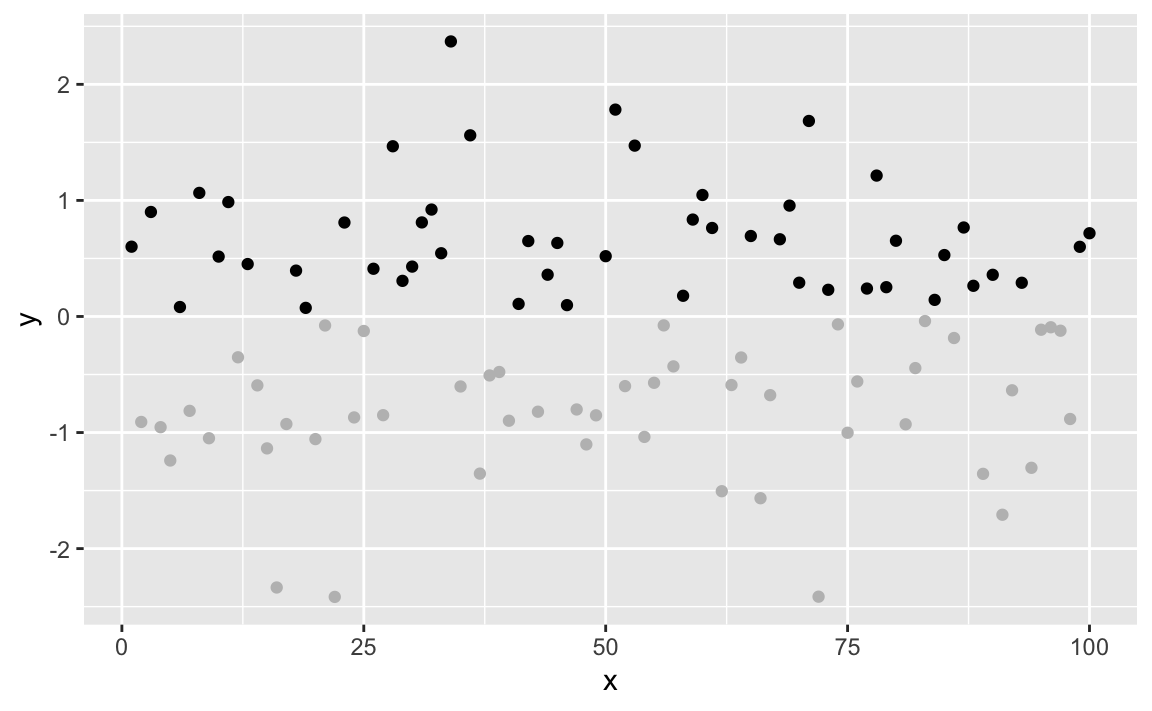
Figure 10-40. Color Shaded Point Plot
The negative values are now plotted in gray because the corresponding
element of colors is "gray".
See Also
See “Understanding the Recycling Rule” regarding the Recycling Rule. Execute colors() to see
a list of available colors, and use geom_segment in ggplot to plot
line segments in multiple colors.
Graphing a Function
Problem
You want to graph the value of a function.
Solution
The ggplot function stat_function will graph a function across a
range. In Figure 10-41 we plot a sin wave across the
range -3 to 3.
ggplot(data.frame(x=c(-3,3)))+aes(x)+stat_function(fun=sin)
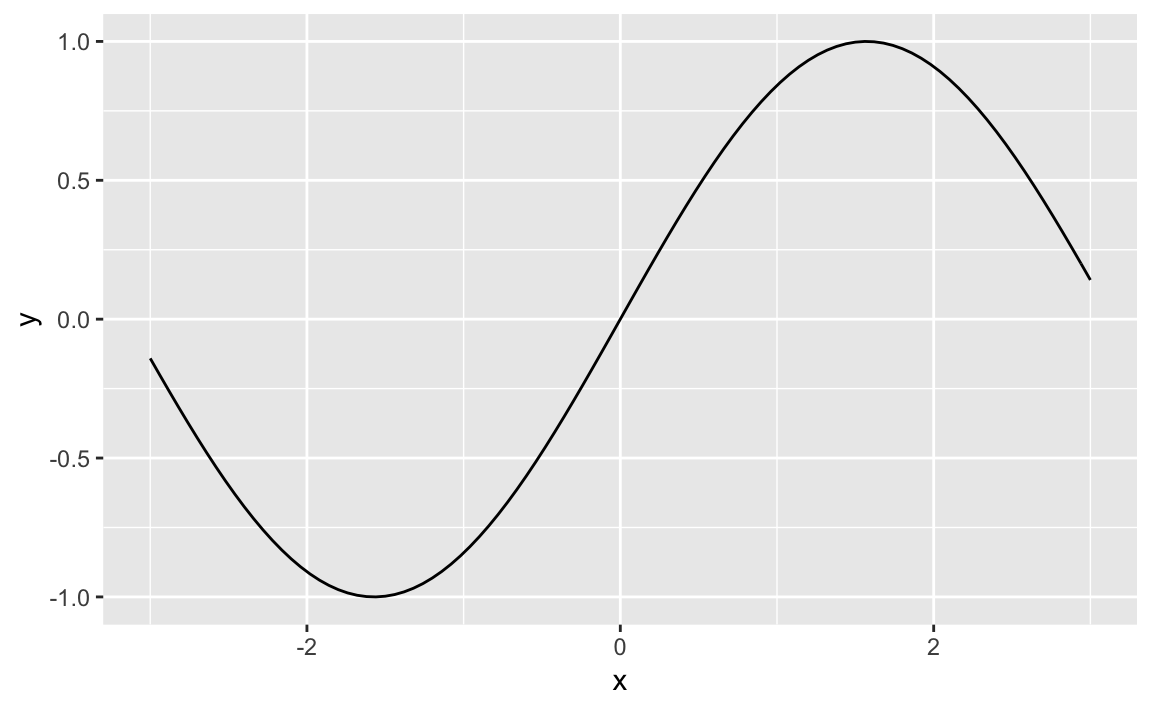
Figure 10-41. Sin Wave Plot
Discussion
It’s pretty common to want to plot a statistical function, such as a
normal distribution across a given range. The stat_function in
ggplot allows us to do thise. We need only supply a data frame with
x value limits and stat_function will calculate the y values, and
plot the results:
ggplot(data.frame(x=c(-3.5,3.5)))+aes(x)+stat_function(fun=dnorm)+ggtitle("Std. Normal Density")
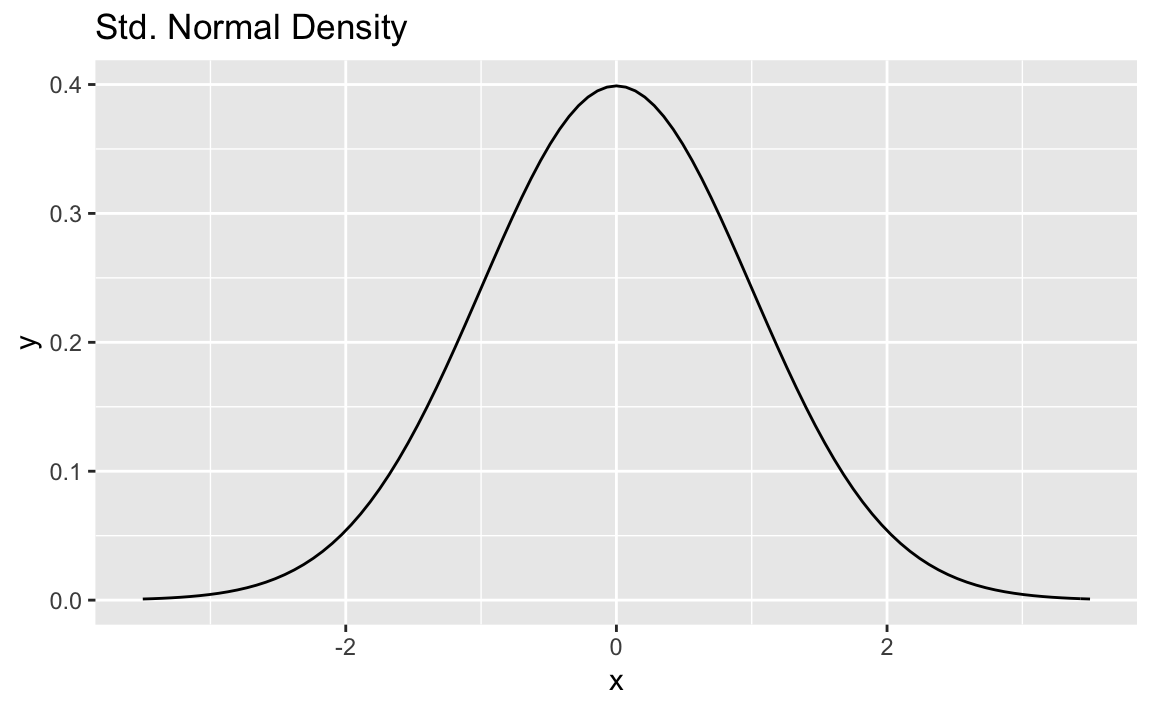
Notice in the chart above we use ggtitle to set the title. If setting
multiple text elements in a ggplot we use labs but when just adding
a title, ggtitle is more concise than
labs(title='Std. Normal Density') although they accomplish the same
thing. See ?labs for more discussion of labels with ggplot
stat_function can graph any function that takes one argument and
returns one value. Let’s create a function and then plot it. Our
function is a dampened sin wave that is a sin wave that loses amplitude
as it moves away from 0:
f<-function(x)exp(-abs(x))*sin(2*pi*x)
ggplot(data.frame(x=c(-3.5,3.5)))+aes(x)+stat_function(fun=f)+ggtitle("Dampened Sine Wave")
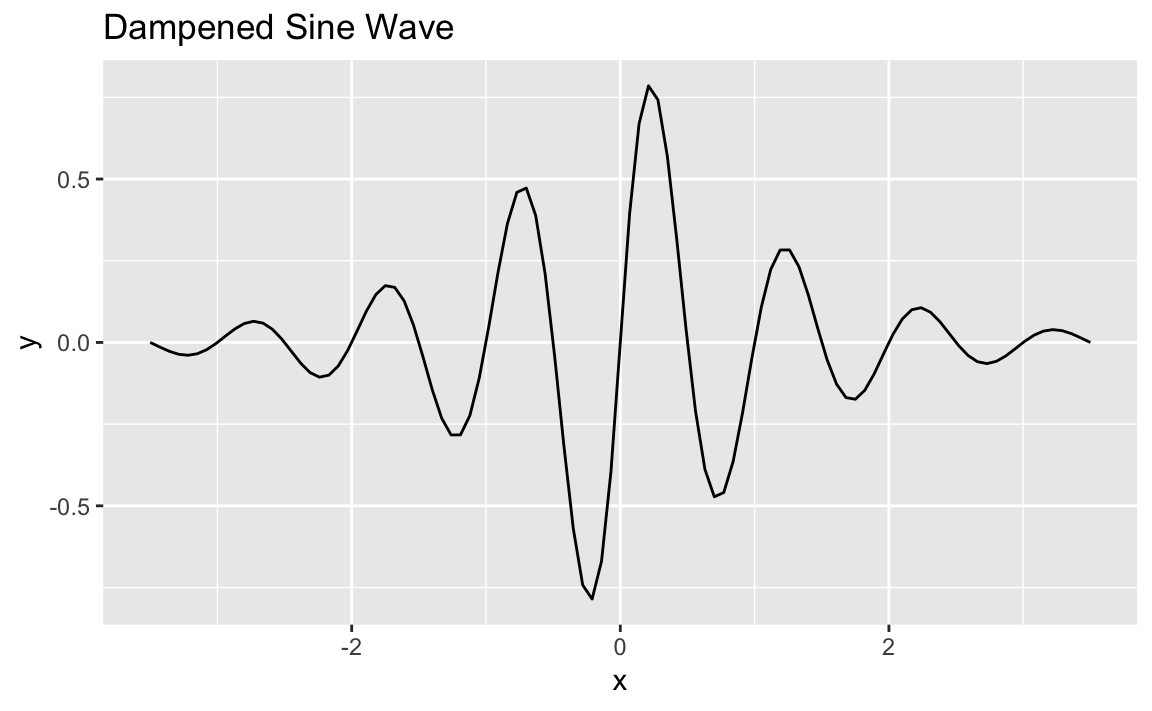
See Also
See Recipe X-X for how to define a function.
Pausing Between Plots
Problem
You are creating several plots, and each plot is overwriting the previous one. You want R to pause between plots so you can view each one before it’s overwritten.
Solution
There is a global graphics option called ask. Set it to TRUE, and R
will pause before each new plot. We turn on this option by passing it to
the par function which sets parameters:
par(ask=TRUE)
When you are tired of R pausing between plots, set it to FALSE:
par(ask=FALSE)
Discussion
When ask is TRUE, R will print this message immediately before
starting a new plot:
Hit <Return> to see next plot:
When you are ready, hit the return or enter key and R will begin the next plot.
This is a Base R Graphics function but you can use it in ggplot if you
wrap your plot function in a print statement in order to get prompted.
Below is an example of a loop that prints a random set of points 5
times. If you run this loop in RStudio, you will be prompted between
each graphic. Notice how we wrap g inside a print call:
par(ask=TRUE)for(iin(11:15)){g<-ggplot(data.frame(x=rnorm(i),y=1:i))+aes(x,y)+geom_point()(g)}# don't forget to turn ask off after you're donepar(ask=FALSE)
See Also
If one graph is overwriting another, consider using “Displaying Several Figures on One Page” to plot multiple graphs in one frame. See Recipe X-X for more about changing graphical parameters.
JDL EDIT MARK
Displaying Several Figures on One Page
Problem
You want to display several plots side by side on one page.
Solution
# example dataz<-rnorm(1000)y<-runif(1000)# plot elementsp1<-ggplot()+geom_point(aes(x=1:1000,y=z))p2<-ggplot()+geom_point(aes(x=1:1000,y=y))p3<-ggplot()+geom_density(aes(z))p4<-ggplot()+geom_density(aes(y))
There are a number of ways to put ggplot graphics into a grid, but one
of the easiest to use and understand is patchwork by Thomas Lin
Pedersen. When this book was written, patchwork was not availiable on
CRAN, but can be installed using devtools:
devtools::install_github("thomasp85/patchwork")
After installing the package we can use it to plot mulitple ggplot
objects using a + between the objects then a call to plot_layout to
arrange the images into a grid as shown in Figure 10-42:
library(patchwork)p1+p2+p3+p4
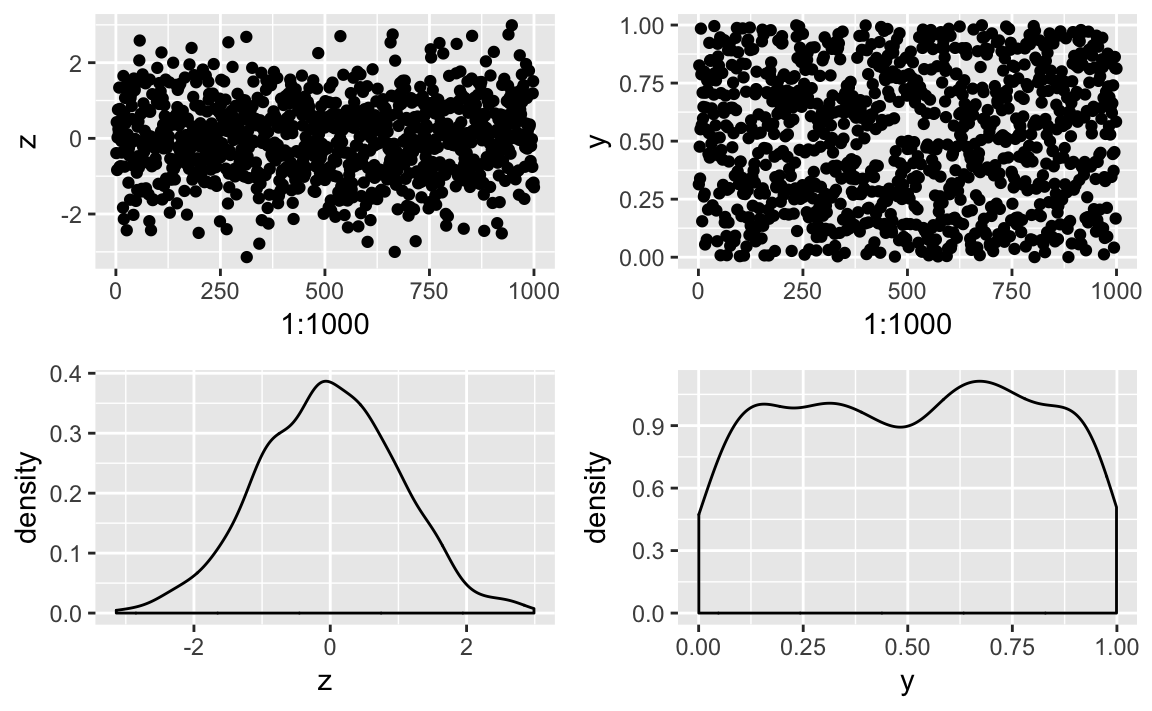
Figure 10-42. A Patchwork Plot
patchwork supports grouping with parenthesis and using / to put
groupings under other elements as illustrated in Figure 10-43.
p3/(p1+p2+p4)
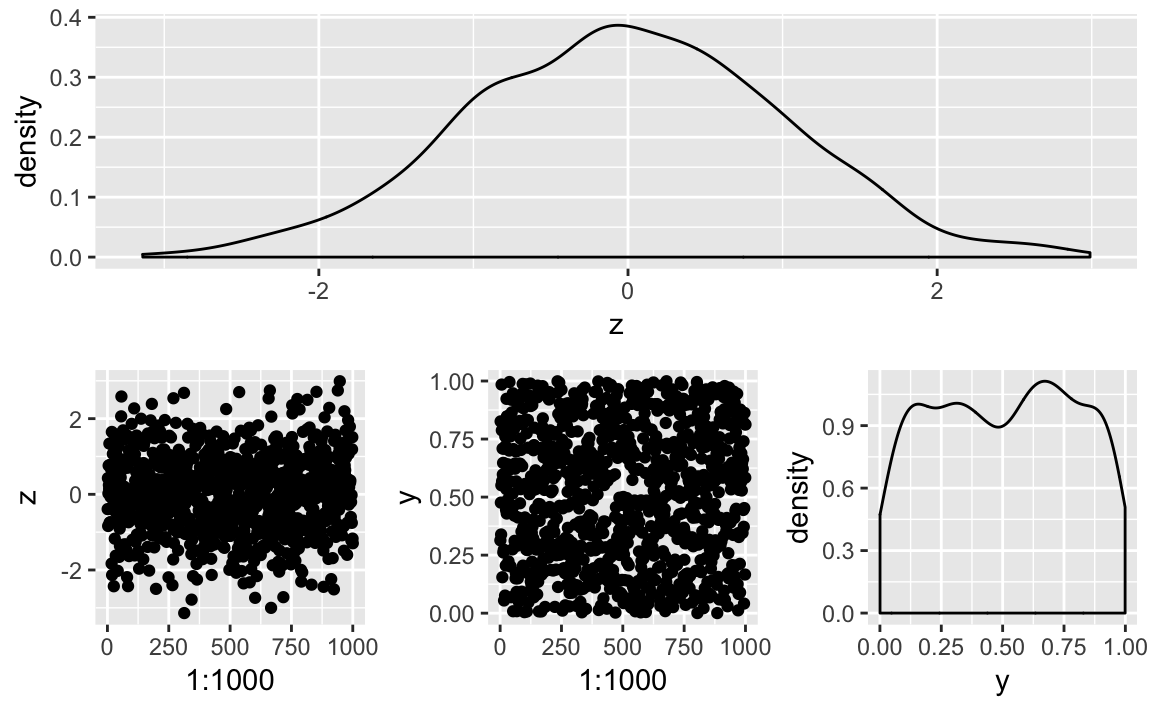
Figure 10-43. A Patchwork 1 / 2 Plot
Discussion
Let’s use a multifigure plot to display four different beta
distributions. Using ggplot and the patchwork package, we can create
a 2 x 2 layout effect by greating four graphics objects then print them
using the + notation from Patchwork:
library(patchwork)df<-data.frame(x=c(0,1))g1<-ggplot(df)+aes(x)+stat_function(fun=function(x)dbeta(x,2,4))+ggtitle("First")g2<-ggplot(df)+aes(x)+stat_function(fun=function(x)dbeta(x,4,1))+ggtitle("Second")g3<-ggplot(df)+aes(x)+stat_function(fun=function(x)dbeta(x,1,1))+ggtitle("Third")g4<-ggplot(df)+aes(x)+stat_function(fun=function(x)dbeta(x,.5,.5))+ggtitle("Fourth")g1+g2+g3+g4+plot_layout(ncol=2,byrow=TRUE)
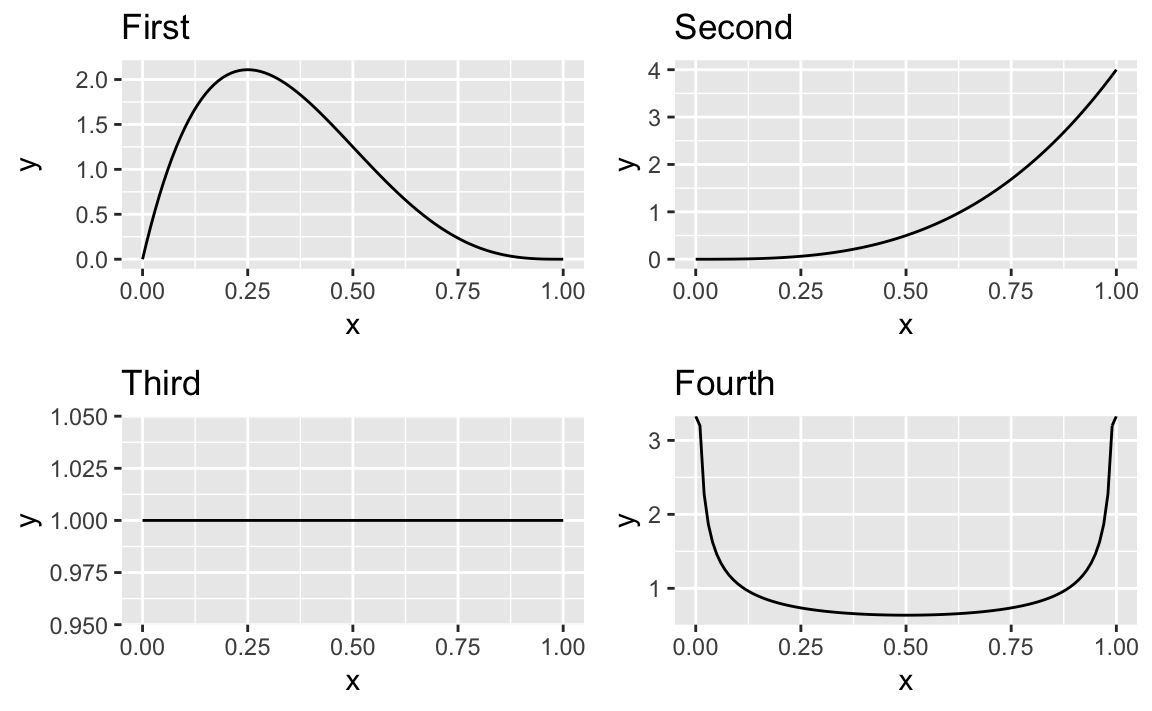
To lay the images out in columns order we could pass the byrow=FALSE
to plot_layout:
g1+g2+g3+g4+plot_layout(ncol=2,byrow=FALSE)
See Also
“Plotting a Density Function” discusses plotting of density functions as we do above.
The grid package and the lattice package contain additional tools
for multifigure layouts with Base Graphics.
Writing Your Plot to a File
Problem
You want to save your graphics in a file, such as a PNG, JPEG, or PostScript file.
Solution
With ggplot figures we can use ggsave to save a displayed image to a
file. ggsave will make some default assumptions about size and file
type for you, allowing you to only specify a filename:
ggsave("filename.jpg")
The file type is derived from the extension you use in the filename you
pass to ggsave. You can control details of size, filetype, and scale
by passing parameters to ggsave. See ?ggsave for specific details.
Discussion
In RStudio, a shortcut is to click on Export in the Plots window and
then click on Save as Image, Save as PDF, or Copy to Clipboard.
The save options will prompt you for a file type and a file name before
writing the file. The Copy to Clipboard option can be handy if you are
manually copying and pasting your graphics into a presentation or word
processor.
Remember that the file will be written to your current working directory
(unless you use an absolute file path), so be certain you know which
directory is your working directory before calling savePlot.
In a non-interactive script using ggplot you can pass plot objects
directly to ggsave so they need not be displayed before saving. In the
prior recipe we created a plot object called g1. We can save it to a
file like this:
ggsave("g1.png",plot=g1,units="in",width=5,height=4)
Note that the units for height and width in ggsave are specified
with the units parameter. In this case we used in for inches, but
ggsave also supports mm and cm for the more metricly inclined.
See Also
See “Getting and Setting the Working Directory” for more about the current working directory.
