Chapter 1. Getting Started and Getting Help
Introduction
This chapter sets the groundwork for the other chapters. It explains how to download, install, and run R.
More importantly, it also explains how to get answers to your questions. The R community provides a wealth of documentation and help. You are not alone. Here are some common sources of help:
- Local, installed documentation
-
When you install R on your computer, a mass of documentation is also installed. You can browse the local documentation (“Viewing the Supplied Documentation”) and search it (“Searching the Supplied Documentation”). We are amazed how often we search the Web for an answer only to discover it was already available in the installed documentation.
- Task views: (http://cran.r-project.org/web/views)
-
A task view describes packages that are specific to one area of statistical work, such as econometrics, medical imaging, psychometrics, or spatial statistics. Each task view is written and maintained by an expert in the field. There are more than 35 such task views, so there is likely to be one or more for your areas of interest. We recommend that every beginner find and read at least one task view in order to gain a sense of R’s possibilities (“Finding Relevant Functions and Packages”).
- Package documentation
-
Most packages include useful documentation. Many also include overviews and tutorials, called “vignettes” in the R community. The documentation is kept with the packages in package repositories, such as CRAN (http://cran.r-project.org/), and it is automatically installed on your machine when you install a package.
- Question and answer (Q&A) websites
-
On a Q&A site, anyone can post a question, and knowledgeable people can respond. Readers vote on the answers, so the best answers tend to emerge over time. All this information is tagged and archived for searching. These sites are a cross between a mailing list and a social network; “Stack Overflow” (http://stackoverflow.com/) is the canonical example.
- The Web
-
The Web is loaded with information about R, and there are R-specific tools for searching it (“Searching the Web for Help”). The Web is a moving target, so be on the lookout for new, improved ways to organize and search information regarding R.
- Mailing lists
-
Volunteers have generously donated many hours of time to answer beginners’ questions that are posted to the R mailing lists. The lists are archived, so you can search the archives for answers to your questions (“Searching the Mailing Lists”).
Downloading and Installing R
Problem
You want to install R on your computer.
Solution
Windows and OS X users can download R from CRAN, the Comprehensive R Archive Network. Linux and Unix users can install R packages using their package management tool:
Windows
-
Open http://www.r-project.org/ in your browser.
-
Click on “CRAN”. You’ll see a list of mirror sites, organized by country.
-
Select a site near you or the top one listed as “0-Cloud” which tends to work well for most locations (https://cloud.r-project.org/)
-
Click on “Download R for Windows” under “Download and Install R”.
-
Click on “base”.
-
Click on the link for downloading the latest version of R (an
.exefile). -
When the download completes, double-click on the
.exefile and answer the usual questions.
OS X
-
Open http://www.r-project.org/ in your browser.
-
Click on “CRAN”. You’ll see a list of mirror sites, organized by country.
-
Select a site near you or the top one listed as “0-Cloud” which tends to work well for most locations.
-
Click on “Download R for (Mac) OS X”.
-
Click on the
.pkgfile for the latest version of R, under “Latest release:”, to download it. -
When the download completes, double-click on the
.pkgfile and answer the usual questions.
Linux or Unix
The major Linux distributions have packages for installing R. Here are some examples:
| Distribution | Package name |
|---|---|
Ubuntu or Debian |
r-base |
Red Hat or Fedora |
R.i386 |
Suse |
R-base |
Use the system’s package manager to download and install the package.
Normally, you will need the root password or sudo privileges;
otherwise, ask a system administrator to perform the installation.
Discussion
Installing R on Windows or OS X is straightforward because there are prebuilt binaries (compiled programs) for those platforms. You need only follow the preceding instructions. The CRAN Web pages also contain links to installation-related resources, such as frequently asked questions (FAQs) and tips for special situations (“Does R run under Windows Vista/7/8/Server 2008?”) that you may find useful.
The best way to install R on Linux or Unix is by using your Linux distribution package manager to install R as a package. The distribution packages greatly streamline both the initial installation and subsequent updates.
On Ubuntu or Debian, use apt-get to download and install R. Run under
sudo to have the necessary privileges:
$sudoapt-getinstallr-base
On Red Hat or Fedora, use yum:
$sudoyuminstallR.i386
Most Linux platforms also have graphical package managers, which you might find more convenient.
Beyond the base packages, we recommend installing the documentation
packages, too. We like to install r-base-html (because we like
browsing the hyperlinked documentation) as well as r-doc-html, which
installs the important R manuals locally:
$sudoapt-getinstallr-base-htmlr-doc-html
Some Linux repositories also include prebuilt copies of R packages available on CRAN. We don’t use them because we’d rather get software directly from CRAN itself, which usually has the freshest versions.
In rare cases, you may need to build R from scratch. You might have an
obscure, unsupported version of Unix; or you might have special
considerations regarding performance or configuration. The build
procedure on Linux or Unix is quite standard. Download the tarball from
the home page of your CRAN mirror; it’s called something like
R-3.5.1.tar.gz, except the “3.5.1” will be replaced by the latest
version. Unpack the tarball, look for a file called INSTALL, and
follow the directions.
See Also
R in a Nutshell (http://oreilly.com/catalog/9780596801717) (O’Reilly) contains more details of downloading and installing R, including instructions for building the Windows and OS X versions. Perhaps the ultimate guide is the one entitled “R Installation and Administration” (http://cran.r-project.org/doc/manuals/R-admin.html), available on CRAN, which describes building and installing R on a variety of platforms.
This recipe is about installing the base package. See “Installing Packages from CRAN” for installing add-on packages from CRAN.
Installing R Studio
Problem
You want a more comprehensive Integrated Development Environment (IDE) than the R default. In other words, you want to install R Studio Desktop.
Solution
Over the past few years R Studio has become the most widly used IDE for
R. We are of the opinion that most all R work should be done in the R
Studio Desktop IDE unless there is a compelling reason to do otherwise.
R Studio makes multiple products including R Studio Desktop, R Studio
Server, R Studio Shiny Server, just to name a few. For this book we will
use the term R Studio to mean R Studio Desktop though most concepts
apply to R Studio Server as well.
To install R Studio, download the latest installer for your platform from the R Studio website: https://www.rstudio.com/products/rstudio/download/
The R Studio Desktop Open Source License version is free to download and use.
Discussion
This book was written and built using R Studio version 1.2.x and R versions 3.5.x. New versions of R Studio are released every few months, so be sure and update regularly. Note that R Studio works with whichever version of R you have installed. So updating to the latest version of R Studio does not upgrade your version of R. R must be upgraded seperatly.
Interacting with R is slightly different in R Studio than in the built in R user interface. For this book, we’ve elected to use R Studio for all examples.
Starting R Studio
Problem
You want to run R Studio on your computer.
Solution
A common point of confusion for new users of R and R Studio is to
accidentally start R when they intended to start R Studio. The easiest
way to ensure you’re actually starting R Studio is to search for
RStudio on your desktop OS. Then use whatever method your OS provides
for pinning the icon somewhere easy to find later.
- Windows
-
Click on the Start Screen menue in the lower left corner of the screen. In the search box, type
RStudio. - OS X
-
Look in your launchpad for the R Studio app or press
command
spaceand typeRstudioto search using Spotlight Search. - Ubuntu
-
Press
Alt+F1and typeRStudioto search for R Studio.
Discussion
Confusion between R and R Studio can easily happen becuase as you can see in Figure 1-1, the icons look similiar.

Figure 1-1. R and R Studio icons in OSX
If you click on the R icon you’ll be greeted by something like Figure
Figure 1-2 which is the Base R interface on a Mac, but certainly
not R Studio.
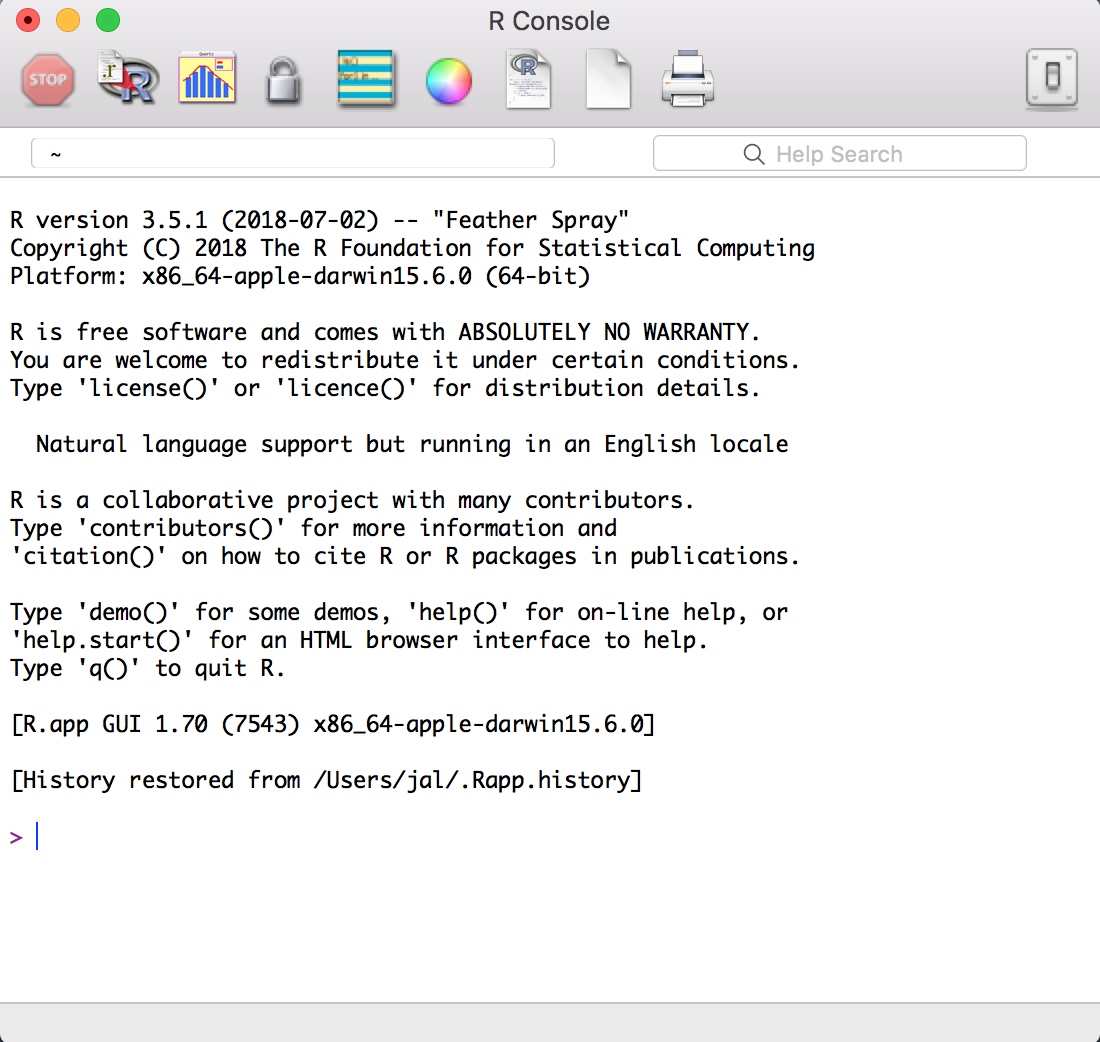
Figure 1-2. The R Console in OSX
When you start R Studio, the default behavior is that R Studio will reopen the last project you were working on in R Studio.
Entering Commands
Problem
You’ve started R Studio. Now what?
Solution
When you start R Studio, the main window on the left is an R session. From there you can enter commands interactivly directly to R.
Discussion
R prompts you with “>”. To get started, just treat R like a big
calculator: enter an expression, and R will evaluate the expression and
print the result:
1+1#> [1] 2
The computer adds one and one, giving two, and displays the result.
The [1] before the 2 might be confusing. To R, the result is a
vector, even though it has only one element. R labels the value with
[1] to signify that this is the first element of the vector… which
is not surprising, since it’s the only element of the vector.
R will prompt you for input until you type a complete expression. The
expression max(1,3,5) is a complete expression, so R stops reading
input and evaluates what it’s got:
max(1,3,5)#> [1] 5
In contrast, “max(1,3,” is an incomplete expression, so R prompts you
for more input. The prompt changes from greater-than (>) to plus
(+), letting you know that R expects more:
max(1,3,+5)#> [1] 5
It’s easy to mistype commands, and retyping them is tedious and frustrating. So R includes command-line editing to make life easier. It defines single keystrokes that let you easily recall, correct, and reexecute your commands. My own typical command-line interaction goes like this:
-
I enter an R expression with a typo.
-
R complains about my mistake.
-
I press the up-arrow key to recall my mistaken line.
-
I use the left and right arrow keys to move the cursor back to the error.
-
I use the Delete key to delete the offending characters.
-
I type the corrected characters, which inserts them into the command line.
-
I press Enter to reexecute the corrected command.
That’s just the basics. R supports the usual keystrokes for recalling and editing command lines, as listed in table @ref(tab:keystrokes).
| Labeled key | Ctrl-key combination | Effect |
|---|---|---|
Up arrow |
Ctrl-P |
Recall previous command by moving backward through the history of commands. |
Down arrow |
Ctrl-N |
Move forward through the history of commands. |
Backspace |
Ctrl-H |
Delete the character to the left of cursor. |
Delete (Del) |
Ctrl-D |
Delete the character to the right of cursor. |
Home |
Ctrl-A |
Move cursor to the start of the line. |
End |
Ctrl-E |
Move cursor to the end of the line. |
Right arrow |
Ctrl-F |
Move cursor right (forward) one character. |
Left arrow |
Ctrl-B |
Move cursor left (back) one character. |
Ctrl-K |
Delete everything from the cursor position to the end of the line. |
|
Ctrl-U |
Clear the whole darn line and start over. |
|
Tab |
Name completion (on some platforms). |
: Keystrokes for command-line editing
On Windows and OS X, you can also use the mouse to highlight commands and then use the usual copy and paste commands to paste text into a new command line.
See Also
See “Typing Less and Accomplishing More”. From the Windows main menu, follow Help →
Console for a complete list of keystrokes useful for command-line
editing.
Exiting from R Studio
Problem
You want to exit from R Studio.
Solution
- Windows
-
Select
File→Quit Sessionfrom the main menu; or click on the X in the upper-right corner of the window frame. - OS X
-
Press CMD-q (apple-q); or click on the red X in the upper-left corner of the window frame.
- Linux or Unix
-
At the command prompt, press Ctrl-D.
On all platforms, you can also use the q function (as in _q_uit) to
terminate the program.
q()
Note the empty parentheses, which are necessary to call the function.
Discussion
Whenever you exit, R typically asks if you want to save your workspace. You have three choices:
-
Save your workspace and exit.
-
Don’t save your workspace, but exit anyway.
-
Cancel, returning to the command prompt rather than exiting.
If you save your workspace, then R writes it to a file called .RData
in the current working directory. Savign the workspace saves any R
objects which you have created. Next time you start R in the same
directory the workspace will automatically load. Saving your workspace
will overwrite the previously saved workspace, if any, so don’t save if
you don’t like the changes to your workspace (e.g., if you have
accidentally erased critical data).
We recommend never saving your workspace when you exit, and instead
always explicitly saving your project, scripts, and data. We also
recommend that you turn off the prompt to save and auto restore of
workspace in R Studio using the Global Options found in the menu Tools
→ Global Options and shown in Figure 1-3. This way
when you exit R and R Studio you will not be prompted to save your
workspace. But keep in mind that any objects created but not saved to
disk will be lost.
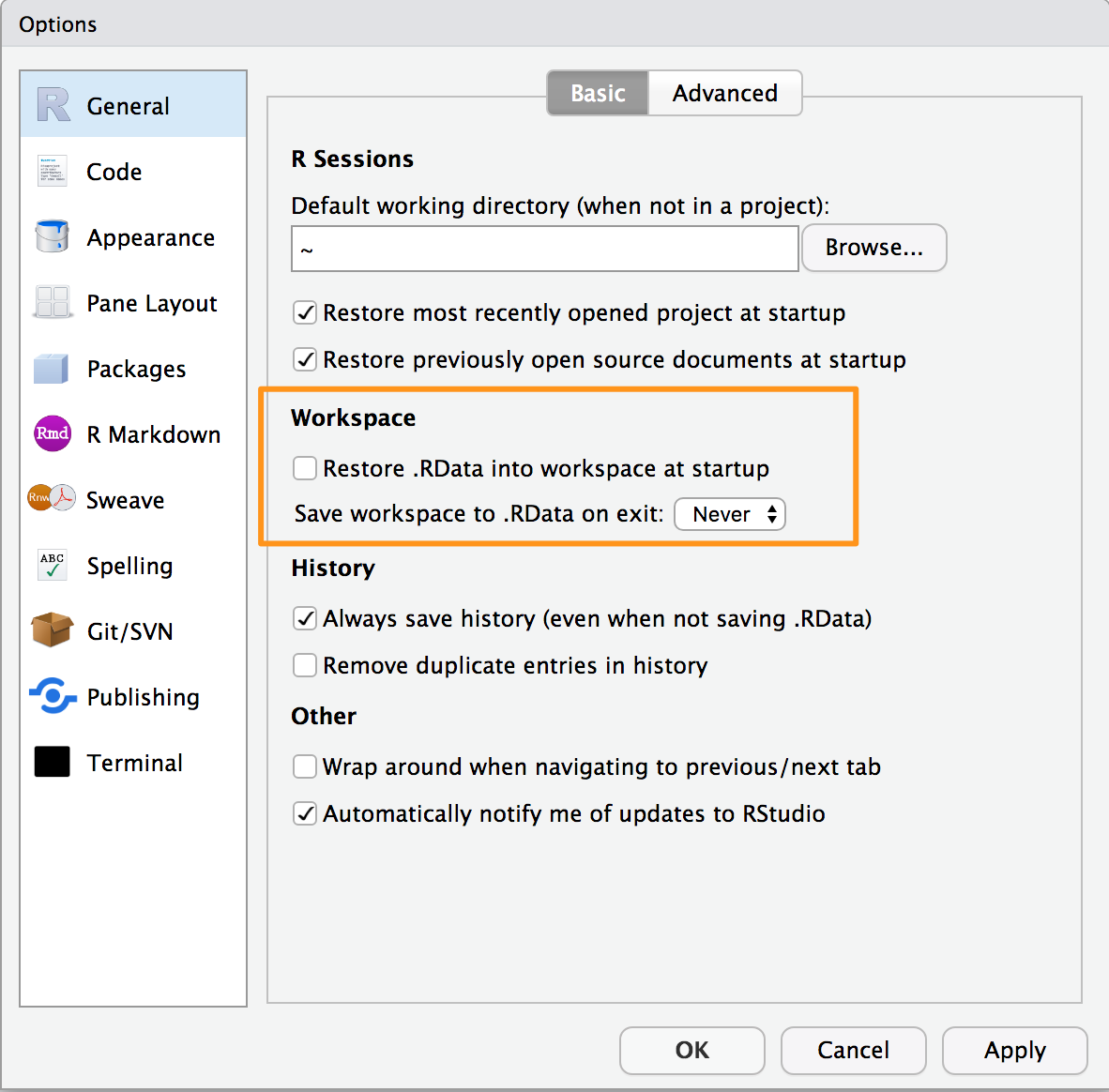
Figure 1-3. Save Workspace Options
See Also
See “Getting and Setting the Working Directory” for more about the current working directory and “Saving Your Workspace” for more about saving your workspace. See Chapter 2 of R in a Nutshell (http://oreilly.com/catalog/9780596801717).
Interrupting R
Problem
You want to interrupt a long-running computation and return to the command prompt without exiting R Studio.
Solution
Press the Esc key on your keyboard, or click on the Session Menu in
R Studio and select Interrupt R
Discussion
Interrupting R means telling R to stop running the current command but without deleting variables from memory or completly closing R Studio. Although, interrupting R can leave your variables in an indeterminate state, depending upon how far the computation had progressed. Check your workspace after interrupting.
See Also
Viewing the Supplied Documentation
Problem
You want to read the documentation supplied with R.
Solution
Use the help.start function to see the documentation’s table of
contents:
help.start()
From there, links are available to all the installed documentation. In R Studio the help will show up in the help pane which by default is on the right hand side of the screen.
In R Studio you can also click help → R Help to get a listng with
help options for both R and R Studio.
Discussion
The base distribution of R includes a wealth of documentation—literally thousands of pages. When you install additional packages, those packages contain documentation that is also installed on your machine.
It is easy to browse this documentation via the help.start function,
which opens on the top-level table of contents. Figure
Figure 1-4 shows how help.start() appears inside the help
pane in R Studio.
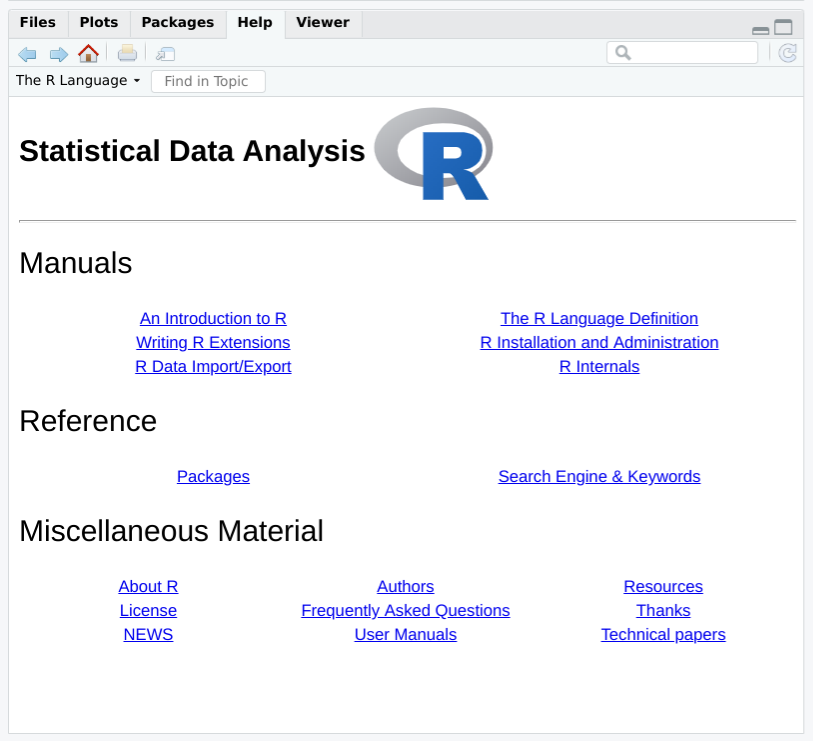
Figure 1-4. R Studio help.start
The two links in the Base R Reference section are especially useful:
- Packages
-
Click here to see a list of all the installed packages, both in the base packages and the additional, installed packages. Click on a package name to see a list of its functions and datasets.
- Search Engine & Keywords
-
Click here to access a simple search engine, which allows you to search the documentation by keyword or phrase. There is also a list of common keywords, organized by topic; click one to see the associated pages.
The Base R documentation shown by typing help.start() is loaded on
your computer when you install R. The R Studio help which you get by
using the menu option help → R Help presents a page with links to R
Studio’s web site. So you will need Internet access to access the R
Studio help links.
See Also
The local documentation is copied from the R Project website, which may have updated documents.
Getting Help on a Function
Problem
You want to know more about a function that is installed on your machine.
Solution
Use help to display the documentation for the function:
help(functionname)
Use args for a quick reminder of the function arguments:
args(functionname)
Use example to see examples of using the function:
example(functionname)
Discussion
We present many R functions in this book. Every R function has more bells and whistles than we can possibly describe. If a function catches your interest, we strongly suggest reading the help page for that function. One of its bells or whistles might be very useful to you.
Suppose you want to know more about the mean function. Use the help
function like this:
help(mean)
This will open the help page for the mean function in the help pane in R
Studio. A shortcut for the help command is to simply type ? followed
by the function name:
?mean
Sometimes you just want a quick reminder of the arguments to a function:
What are they, and in what order do they occur? Use the args function:
args(mean)#> function (x, ...)#> NULL
args(sd)#> function (x, na.rm = FALSE)#> NULL
The first line of output from args is a synopsis of the function call.
For mean, the synopsis shows one argument, x, which is a vector of
numbers. For sd, the synopsis shows the same vector, x, and an
optional argument called na.rm. (You can ignore the second line of
output, which is often just NULL.) In R Studio you will see the args
output as a floating tool tip over your cursor when you type a function
name as shown in figure Figure 1-5.
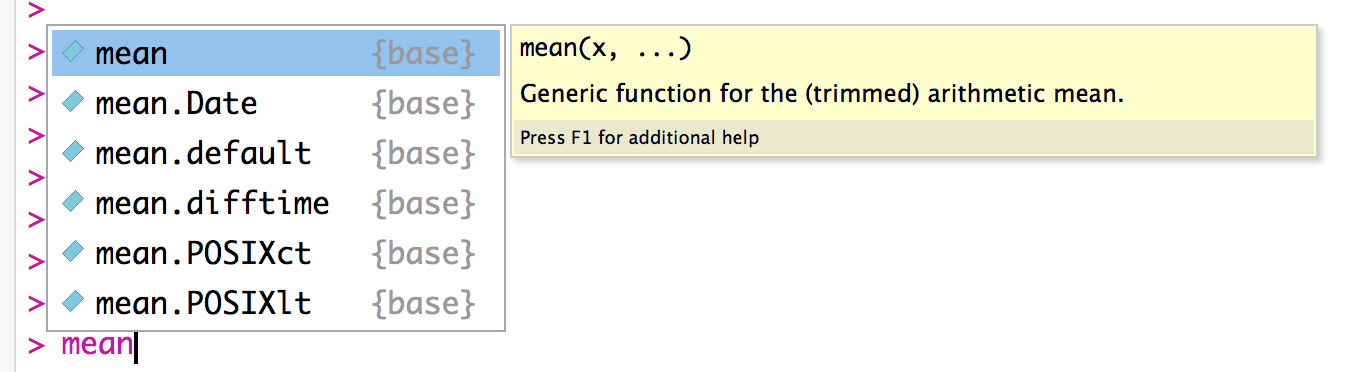
Figure 1-5. R Studio Tooltip
Most documentation for functions includes example code near the end of
the document. A cool feature of R is that you can request that it
execute the examples, giving you a little demonstration of the
function’s capabilities. The documentation for the mean function, for
instance, contains examples, but you don’t need to type them yourself.
Just use the example function to watch them run:
example(mean)#>#> mean> x <- c(0:10, 50)#>#> mean> xm <- mean(x)#>#> mean> c(xm, mean(x, trim = 0.10))#> [1] 8.75 5.50
The user typed example(mean). Everything else was produced by R, which
executed the examples from the help page and displayed the results.
See Also
See “Searching the Supplied Documentation” for searching for functions and “Displaying Loaded Packages via the Search Path” for more about the search path.
Searching the Supplied Documentation
Problem
You want to know more about a function that is installed on your
machine, but the help function reports that it cannot find
documentation for any such function.
Alternatively, you want to search the installed documentation for a keyword.
Solution
Use help.search to search the R documentation on your computer:
help.search("pattern")
A typical pattern is a function name or keyword. Notice that it must be enclosed in quotation marks.
For your convenience, you can also invoke a search by using two question marks (in which case the quotes are not required). Note that searching for a function by name uses one question mark while searching for a text pattern uses two:
>??pattern
Discussion
You may occasionally request help on a function only to be told R knows nothing about it:
help(adf.test)#> No documentation for 'adf.test' in specified packages and libraries:#> you could try '??adf.test'
This can be frustrating if you know the function is installed on your machine. Here the problem is that the function’s package is not currently loaded, and you don’t know which package contains the function. It’s a kind of catch-22 (the error message indicates the package is not currently in your search path, so R cannot find the help file; see “Displaying Loaded Packages via the Search Path” for more details).
The solution is to search all your installed packages for the function.
Just use the help.search function, as suggested in the error message:
help.search("adf.test")
The search will produce a listing of all packages that contain the function:
Helpfileswithaliasorconceptortitlematching'adf.test'usingregularexpressionmatching:tseries::adf.testAugmentedDickey-FullerTestType'?PKG::FOO'toinspectentry'PKG::FOO TITLE'.
The output above indicates that the tseries package contains the
adf.test function. You can see its documentation by explicitly telling
help which package contains the function:
help(adf.test,package="tseries")
or you can use the double colon operator to tell R to look in a specific package:
?tseries::adf.test
You can broaden your search by using keywords. R will then find any installed documentation that contains the keywords. Suppose you want to find all functions that mention the Augmented Dickey–Fuller (ADF) test. You could search on a likely pattern:
help.search("dickey-fuller")
On my machine, the result looks like this because I’ve installed two
additional packages (fUnitRoots and urca) that implement the ADF
test:
Helpfileswithaliasorconceptortitlematching'dickey-fuller'usingfuzzymatching:fUnitRoots::DickeyFullerPValuesDickey-FullerpValuestseries::adf.testAugmentedDickey-FullerTesturca::ur.dfAugmented-Dickey-FullerUnitRootTestType'?PKG::FOO'toinspectentry'PKG::FOO TITLE'.
See Also
You can also access the local search engine through the documentation browser; see “Viewing the Supplied Documentation” for how this is done. See “Displaying Loaded Packages via the Search Path” for more about the search path and “Listing Files” for getting help on functions.
Getting Help on a Package
Problem
You want to learn more about a package installed on your computer.
Solution
Use the help function and specify a package name (without a function
name):
help(package="packagename")
Discussion
Sometimes you want to know the contents of a package (the functions and datasets). This is especially true after you download and install a new package, for example. The help function can provide the contents plus other information once you specify the package name.
This call to help will display the information for the tseries
package, a standard package in the base distribution:
help(package="tseries")
The information begins with a description and continues with an index of functions and datasets. In R Studio, the HTML formatted help page will open in the help window of the IDE.
Some packages also include vignettes, which are additional documents such as introductions, tutorials, or reference cards. They are installed on your computer as part of the package documentation when you install the package. The help page for a package includes a list of its vignettes near the bottom.
You can see a list of all vignettes on your computer by using the
vignette function:
vignette()
In R Studio this will open a new tab and list every package installed on your computer which includes vignettes and a list of vignette names and descriptions.
You can see the vignettes for a particular package by including its name:
vignette(package="packagename")
Each vignette has a name, which you use to view the vignette:
vignette("vignettename")
See Also
See “Getting Help on a Function” for getting help on a particular function in a package.
Searching the Web for Help
Problem
You want to search the Web for information and answers regarding R.
Solution
Inside R, use the RSiteSearch function to search by keyword or phrase:
RSiteSearch("key phrase")
Inside your browser, try using these sites for searching:
- RSeek: http://rseek.org
-
This is a Google custom search that is focused on R-specific websites.
- Stack Overflow: http://stackoverflow.com/
-
Stack Overflow is a searchable Q&A site from Stack Exchange oriented toward programming issues such as data structures, coding, and graphics. http://stats.stackexchange.com/[Cross Validated:
- http://stats.stackexchange.com/]
-
Cross Validated is a Stack Exchange site focused on statistics, machine learning, and data analysis rather than programming. Cross Validated is a good place for questions about what statistical method to use.
Discussion
The RSiteSearch function will open a browser window and direct it to
the search engine on the R Project website
(http://search.r-project.org/). There you will see an initial search
that you can refine. For example, this call would start a search for
“canonical correlation”:
RSiteSearch("canonical correlation")
This is quite handy for doing quick web searches without leaving R. However, the search scope is limited to R documentation and the mailing-list archives.
The rseek.org site provides a wider search. Its virtue is that it harnesses the power of the Google search engine while focusing on sites relevant to R. That eliminates the extraneous results of a generic Google search. The beauty of rseek.org is that it organizes the results in a useful way.
Figure Figure 1-6 shows the results of visiting rseek.org and searching for “canonical correlation”. The left side of the page shows general results for search R sites. The right side is a tabbed display that organizes the search results into several categories:
-
Introductions
-
Task Views
-
Support Lists
-
Functions
-
Books
-
Blogs
-
Related Tools
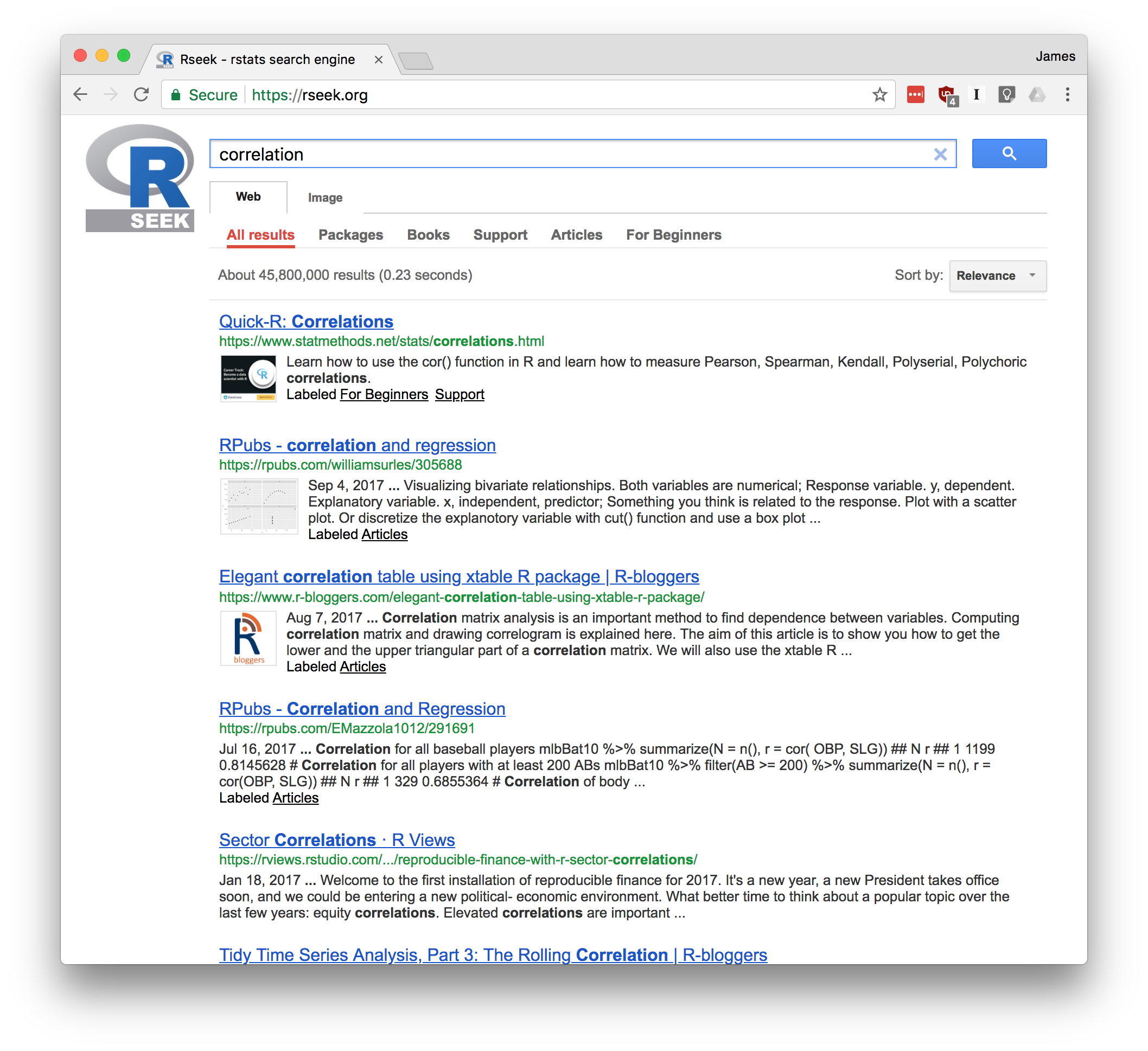
Figure 1-6. RSeek
If you click on the Introductions tab, for example, you’ll find tutorial material. The Task Views tab will show any Task View that mentions your search term. Likewise, clicking on Functions will show links to relevant R functions. This is a good way to zero in on search results.
Stack Overflow (http://stackoverflow.com/) is a Q&A site, which means that anyone can submit a question and experienced users will supply answers—often there are multiple answers to each question. Readers vote on the answers, so good answers tend to rise to the top. This creates a rich database of Q&A dialogs, which you can search. Stack Overflow is strongly problem oriented, and the topics lean toward the programming side of R.
Stack Overflow hosts questions for many programming languages;
therefore, when entering a term into their search box, prefix it with
[r] to focus the search on questions tagged for R. For example,
searching via [r] standard error will select only the questions tagged
for R and will avoid the Python and C++ questions.
Stack Overflow also includes a wiki about the R language that is an excellent community curreated list of online R resources: https://stackoverflow.com/tags/r/info
Stack Exchange (parent company of Stack Overflow) has a Q&A area for statistical analysis called Cross Validated: https://stats.stackexchange.com/. This area is more focused on statistics than programming, so use this site when seeking answers that are more concerned with statistics in general and less with R in particular.
See Also
If your search reveals a useful package, use “Installing Packages from CRAN” to install it on your machine.
Finding Relevant Functions and Packages
Problem
Of the 10,000+ packages for R, you have no idea which ones would be useful to you.
Solution
-
Visit the list of task views at http://cran.r-project.org/web/views/ Find and read the task view for your area, which will give you links to and descriptions of relevant packages. Or visit http://rseek.org, search by keyword, click on the Task Views tab, and select an applicable task view.
-
Visit crantastic (http://crantastic.org/) and search for packages by keyword.
-
To find relevant functions, visit http://rseek.org, search by name or keyword, and click on the Functions tab.
-
To discover packages related to a certain field, explore CRAN Task Views (https://cran.r-project.org/web/views/).
Discussion
This problem is especially vexing for beginners. You think R can solve your problems, but you have no idea which packages and functions would be useful. A common question on the mailing lists is: “Is there a package to solve problem X?” That is the silent scream of someone drowning in R.
As of this writing, there are more than 10,000 packages available for free download from CRAN. Each package has a summary page with a short description and links to the package documentation. Once you’ve located a potentially interesting package, you would typically click on the “Reference manual” link to view the PDF documentation with full details. (The summary page also contains download links for installing the package, but you’ll rarely install the package that way; see “Installing Packages from CRAN”.)
Sometimes you simply have a generic interest—such as Bayesian analysis, econometrics, optimization, or graphics. CRAN contains a set of task view pages describing packages that may be useful. A task view is a great place to start since you get an overview of what’s available. You can see the list of task view pages at CRAN Task Views (http://cran.r-project.org/web/views/) or search for them as described in the Solution. Task Views on CRAN list a number of broad fields and show packages that are used in each field. For example, there are Task Views for high performance computing, genetics, time series, and social science, just to name a few.
Suppose you happen to know the name of a useful package—say, by seeing it mentioned online. A complete, alphabetical list of packages is available at CRAN (http://cran.r-project.org/web/packages/) with links to the package summary pages.
See Also
You can download and install an R package called sos that provides
powerful other ways to search for packages; see the vignette at
SOS
(http://cran.r-project.org/web/packages/sos/vignettes/sos.pdf).
Searching the Mailing Lists
Problem
You have a question, and you want to search the archives of the mailing lists to see whether your question was answered previously.
Solution
-
Open Nabble (http://r.789695.n4.nabble.com/) in your browser. Search for a keyword or other search term from your question. This will show results from the support mailing lists.
Discussion
This recipe is really just an application of “Searching the Web for Help”. But it’s an important application because you should search the mailing list archives before submitting a new question to the list. Your question has probably been answered before.
See Also
CRAN has a list of additional resources for searching the Web; see CRAN Search (http://cran.r-project.org/search.html).
Submitting Questions to Stack Overflow or Elsewhere in the Community
Problem
You have a question you can’t find the answer to online. So you want to submit a question to the R community.
Solution
The first step to asking a question online is to create a reproducable example. Having example code that someone can run and see exactly your problem is to most critical part of asking for help online. A question with a good reproducable example has three componenets:
-
Example Data - This can be simulated data or some real data that you provide
-
Example Code - This code shows what you have tried or an error you are having
-
Written Description - This is where you explain what you have, what you’d like to have and what you have tried that didn’t work.
The details of writing a reproducable example are below in the
discussion. Once you have a reproducable example, you can post your
quesion on Stack Overflow via https://stackoverflow.com/questions/ask.
Be sure and include the r tag in the Tags section of the ask page.
Or if your discussion is more general or related to concepts instead of specific syntax, R Studio runs an R Studio Community discussion forum at https://community.rstudio.com/. Note that the site is broken into multiple topics, so pick the topic category that best fits your question.
Or you may submit your question to the R Mailing lists (but don’t submit to multiple sites, the mailing lists, and Stack Overflow as that’s considered rude cross posting):
The Mailing Lists (http://www.r-project.org/mail.html) page contains general information and instructions for using the R-help mailing list. Here is the general process:
-
Subscribe to the R-help list at the “Main R Mailing List” (https://stat.ethz.ch/mailman/listinfo/r-help).
-
Write your question carefully and correctly and include your reproducable example.
-
Mail your question to r-help@r-project.org.
Discussion
The R mailing list, Stack Overflow, and the R Studio Community site are great resources, but please treat them as a last resort. Read the help pages, read the documentation, search the help list archives, and search the Web. It is most likely that your question has already been answered. Don’t kid yourself: very few questions are unique. If you’ve exhausted all other options, maybe it’s time to create a good question.
The reproducable example is the crux of a good help reqeust. The first
step is example data. A good way to get example data is to simulate the
data using a few R functions. The following example creates a data frame
called example_df that has three columns, each of a different data
type:
set.seed(42)n<-4example_df<-data.frame(some_reals=rnorm(n),some_letters=sample(LETTERS,n,replace=TRUE),some_ints=sample(1:10,n,replace=TRUE))example_df#> some_reals some_letters some_ints#> 1 1.371 R 10#> 2 -0.565 S 3#> 3 0.363 L 5#> 4 0.633 S 10
Note that this example uses the command set.seed() at the beginning.
This ensures that every time this code is run the answers will be the
same. The n value is the number of rows of example data you would like
to create. Make your example data as simple as possible to illustrate
your question.
An alternative to creating simulated data is to use example data that
comes with R. For example, the dataset mtcars contains a data frame
with 32 records about different car models:
data(mtcars)head(mtcars)#> mpg cyl disp hp drat wt qsec vs am gear carb#> Mazda RX4 21.0 6 160 110 3.90 2.62 16.5 0 1 4 4#> Mazda RX4 Wag 21.0 6 160 110 3.90 2.88 17.0 0 1 4 4#> Datsun 710 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1#> Hornet 4 Drive 21.4 6 258 110 3.08 3.21 19.4 1 0 3 1#> Hornet Sportabout 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2#> Valiant 18.1 6 225 105 2.76 3.46 20.2 1 0 3 1
If your example is only reproducable with a bit of your own data, you
can use dput() to put a small bit of your own data in a string which
you can put in your example. We’ll illustrate that using two rows from
the mtcars data:
dput(head(mtcars,2))#> structure(list(mpg = c(21, 21), cyl = c(6, 6), disp = c(160,#> 160), hp = c(110, 110), drat = c(3.9, 3.9), wt = c(2.62, 2.875#> ), qsec = c(16.46, 17.02), vs = c(0, 0), am = c(1, 1), gear = c(4,#> 4), carb = c(4, 4)), row.names = c("Mazda RX4", "Mazda RX4 Wag"#> ), class = "data.frame")
You can put the resulting structure() directly in your question:
example_df<-structure(list(mpg=c(21,21),cyl=c(6,6),disp=c(160,160),hp=c(110,110),drat=c(3.9,3.9),wt=c(2.62,2.875),qsec=c(16.46,17.02),vs=c(0,0),am=c(1,1),gear=c(4,4),carb=c(4,4)),row.names=c("Mazda RX4","Mazda RX4 Wag"),class="data.frame")example_df#> mpg cyl disp hp drat wt qsec vs am gear carb#> Mazda RX4 21 6 160 110 3.9 2.62 16.5 0 1 4 4#> Mazda RX4 Wag 21 6 160 110 3.9 2.88 17.0 0 1 4 4
The second part of a good reproducable example is the example minimal
code. The code example should be as simple as possible and illustrate
what you are trying to do or have already tried. This should not be a
big block of code with many different things going on. Boil your example
down to only the minimal amount of code needed. If you use any packages
be sure and include the library() call at the beginning of your code.
Also, don’t include anything in your question that will harm the state
of someone running your question code, such as rm(list=ls()) which
would delete all R objects in memory. Have empathy for the person trying
to help you and realize that they are volunteering their time to help
you out and may run your code on the same machine they do their own
work.
To test your example, open a new R session and try running your example.
Once you have edited your code, it’s time to give just a bit more
information to your potential question answerer. In the plain text of
the question, describe what you were trying to do, what you’ve tried,
and your question. Be as conscise as possible. Much like with the
example code, your objective is to communicate as efficiently as
possible with the person reading your question. You may find it helpful
to include in your description which version of R you are running as
well as which platform (Windows, Mac, Linux). You can get that
information easily with the sessionInfo() command.
If you are going to submit your question to the R mailing lists, you should know there are actually several mailing lists. R-help is the main list for general questions. There are also many special interest group (SIG) mailing lists dedicated to particular domains such as genetics, finance, R development, and even R jobs. You can see the full list at https://stat.ethz.ch/mailman/listinfo. If your question is specific to one such domain, you’ll get a better answer by selecting the appropriate list. As with R-help, however, carefully search the SIG list archives before submitting your question.
See Also
An excellent essay by Eric Raymond and Rick Moen is entitled “How to Ask Questions the Smart Way” (http://www.catb.org/~esr/faqs/smart-questions.html). We suggest that you read it before submitting any question. Seriously. Read it.
Stack Overflow has an excellent question that includes details about producing a reproducable example. You can find that here: https://stackoverflow.com/q/5963269/37751
Jenny Bryan has a great R package called reprex that helps in the
creation of a good reproduable example and the package has helper
functions that will help you write the markdown text for sites like
Stack Overflow. You can find that package on her Github page:
https://github.com/tidyverse/reprex
