Chapter 14. Trees and Forests
14.0 Introduction
Tree-based learning algorithms are a broad and popular family of related non-parametric, supervised methods for both classification and regression. The basis of tree-based learners is the decision tree wherein a series of decision rules (e.g., “If their gender is male…”) are chained. The result looks vaguely like an upside-down tree, with the first decision rule at the top and subsequent decision rules spreading out below. In a decision tree, every decision rule occurs at a decision node, with the rule creating branches leading to new nodes. A branch without a decision rule at the end is called a leaf.
One reason for the popularity of tree-based models is their interpretability. In fact, decision trees can literally be drawn out in their complete form (see Recipe 14.3) to create a highly intuitive model. From this basic tree system comes a wide variety of extensions from random forests to stacking. In this chapter we will cover how to train, handle, adjust, visualize, and evaluate a number of tree-based models.
14.1 Training a Decision Tree Classifier
Solution
Use scikit-learn’s DecisionTreeClassifier:
# Load librariesfromsklearn.treeimportDecisionTreeClassifierfromsklearnimportdatasets# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create decision tree classifier objectdecisiontree=DecisionTreeClassifier(random_state=0)# Train modelmodel=decisiontree.fit(features,target)
Discussion
Decision tree learners attempt to find a decision rule that produces the
greatest decrease in impurity at a node. While there are a number of
measurements of impurity, by default DecisionTreeClassifier uses Gini
impurity:
where G(t) is the Gini impurity at node t and pi is the proportion of observations of class c at node t. This process of finding the decision rules that create splits to increase impurity is repeated recursively until all leaf nodes are pure (i.e., contain only one class) or some arbitrary cut-off is reached.
In scikit-learn, DecisionTreeClassifier operates like other learning
methods; after the model is trained using fit we can use the
model to predict the class of an observation:
# Make new observationobservation=[[5,4,3,2]]# Predict observation's classmodel.predict(observation)
array([1])
We can also see the predicted class probabilities of the observation:
# View predicted class probabilities for the three classesmodel.predict_proba(observation)
array([[ 0., 1., 0.]])
Finally, if we want to use a different impurity measurement we can use
the criterion parameter:
# Create decision tree classifier object using entropydecisiontree_entropy=DecisionTreeClassifier(criterion='entropy',random_state=0)# Train modelmodel_entropy=decisiontree_entropy.fit(features,target)
See Also
14.2 Training a Decision Tree Regressor
Solution
Use scikit-learn’s DecisionTreeRegressor:
# Load librariesfromsklearn.treeimportDecisionTreeRegressorfromsklearnimportdatasets# Load data with only two featuresboston=datasets.load_boston()features=boston.data[:,0:2]target=boston.target# Create decision tree classifier objectdecisiontree=DecisionTreeRegressor(random_state=0)# Train modelmodel=decisiontree.fit(features,target)
Discussion
Decision tree regression works similarly to decision tree classification; however, instead of reducing Gini impurity or entropy, potential splits are by default measured on how much they reduce mean squared error (MSE):
where yi is the true value of the target and
is the predicted value. In scikit-learn,
decision tree regression can be conducted using DecisionTreeRegressor.
Once we have trained a decision tree, we can use it to predict the
target value for an observation:
# Make new observationobservation=[[0.02,16]]# Predict observation's valuemodel.predict(observation)
array([ 33.])
Just like with DecisionTreeClassifier we can use the criterion
parameter to select the desired measurement of split quality. For
example, we can construct a tree whose splits reduce mean absolute error
(MAE):
# Create decision tree classifier object using entropydecisiontree_mae=DecisionTreeRegressor(criterion="mae",random_state=0)# Train modelmodel_mae=decisiontree_mae.fit(features,target)
14.3 Visualizing a Decision Tree Model
Solution
Export the decision tree model into DOT format, then visualize:
# Load librariesimportpydotplusfromsklearn.treeimportDecisionTreeClassifierfromsklearnimportdatasetsfromIPython.displayimportImagefromsklearnimporttree# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create decision tree classifier objectdecisiontree=DecisionTreeClassifier(random_state=0)# Train modelmodel=decisiontree.fit(features,target)# Create DOT datadot_data=tree.export_graphviz(decisiontree,out_file=None,feature_names=iris.feature_names,class_names=iris.target_names)# Draw graphgraph=pydotplus.graph_from_dot_data(dot_data)# Show graphImage(graph.create_png())
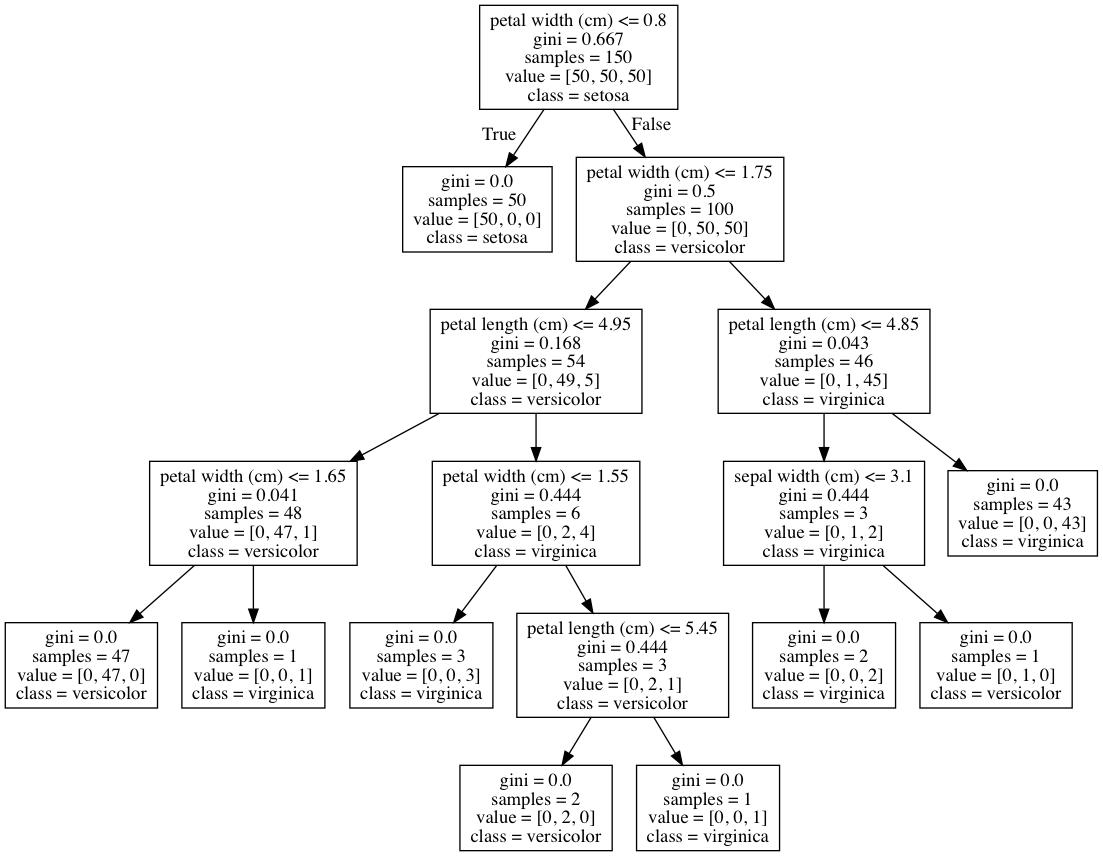
Discussion
One of the advantages of decision tree classifiers is that we can visualize the entire trained model—making decision trees one of the most interpretable models in machine learning. In our solution, we exported our trained model in DOT format (a graph description language) and then used that to draw the graph.
If we look at the root node, we can see the decision rule is that if petal widths are less than or equal to 0.8, then go to the left branch;
if not, go to the right branch. We can also see the Gini impurity index
(0.667), the number of observations (150), the number of observations in each class ([50,50,50]), and the class the observations would be predicted to be if we stopped at that node (setosa). We can also see that at that node the learner found that a single decision rule (petal width (cm) <= 0.8) was able to perfectly identify all of the setosa class observations. Furthermore, with one more decision rule with the same feature (petal width (cm) <= 1.75) the decision tree is able to correctly classify 144 of 150 observations. This makes petal width a very important feature!
If we want to use the decision tree in other applications or reports, we can easily export the visualization into PDF or a PNG image:
# Create PDFgraph.write_pdf("iris.pdf")
True
# Create PNGgraph.write_png("iris.png")
True
While this solution visualized a decision tree classifier, it can just as easily be used to visualize a decision tree regressor.
Note: macOS users might have to install GraphViz’s executable to run the
preceding code. This can be done using Homebrew: brew install graphviz. For Homebrew installation instructions, visit Homebrew’s website.
See Also
14.4 Training a Random Forest Classifier
Solution
Train a random forest classification model using scikit-learn’s
RandomForestClassifier:
# Load librariesfromsklearn.ensembleimportRandomForestClassifierfromsklearnimportdatasets# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create random forest classifier objectrandomforest=RandomForestClassifier(random_state=0,n_jobs=-1)# Train modelmodel=randomforest.fit(features,target)
Discussion
A common problem with decision trees is that they tend to fit the training data too closely (i.e., overfitting). This has motivated the widespread use of an ensemble learning method called random forest. In a random forest, many decision trees are trained, but each tree only receives a bootstrapped sample of observations (i.e., a random sample of observations with replacement that matches the original number of observations) and each node only considers a subset of features when determining the best split. This forest of randomized decision trees (hence the name) votes to determine the predicted class.
As we can see by comparing this solution to Recipe 14.1, scikit-learn’s
RandomForestClassifier works similarly to DecisionTreeClassifier:
# Make new observationobservation=[[5,4,3,2]]# Predict observation's classmodel.predict(observation)
array([1])
RandomForestClassifier also uses many of the same parameters as
DecisionTreeClassifier. For example, we can change the measure of split
quality used:
# Create random forest classifier object using entropyrandomforest_entropy=RandomForestClassifier(criterion="entropy",random_state=0)# Train modelmodel_entropy=randomforest_entropy.fit(features,target)
However, being a forest rather than an individual decision tree,
RandomForestClassifier has certain parameters that are either unique
to random forests or particularly important. First, the max_features parameter determines the maximum number of features to be considered at each node and takes a number of arguments including integers (number of features), floats (percentage of features), and sqrt (square root of the number of features). By default, max_features is set to auto, which acts the same as sqrt. Second, the bootstrap parameter allows us to set whether the subset of observations considered for a tree is created using sampling with replacement (the default setting) or without replacement. Third, n_estimators sets the number of decision trees to include in the forest. In Recipe 10.4 we treated n_estimators as a hyperparameter and visualized the effect of increasing the number of trees on an evaluation metric. Finally, while not specific to random forest classifiers, because we are effectively training many decision tree models, it is often useful to use all available cores by setting n_jobs=-1.
See Also
14.5 Training a Random Forest Regressor
Solution
Train a random forest regression model using scikit-learn’s
RandomForestRegressor:
# Load librariesfromsklearn.ensembleimportRandomForestRegressorfromsklearnimportdatasets# Load data with only two featuresboston=datasets.load_boston()features=boston.data[:,0:2]target=boston.target# Create random forest classifier objectrandomforest=RandomForestRegressor(random_state=0,n_jobs=-1)# Train modelmodel=randomforest.fit(features,target)
Discussion
Just like how we can make a forest of decision tree classifiers, we can
make a forest of decision tree regressors where each tree uses a
bootstrapped subset of observations and at each node the decision rule
considers only a subset of features. As with RandomForestClassifier we
have certain important parameters:
See Also
14.6 Identifying Important Features in Random Forests
Solution
Calculate and visualize the importance of each feature:
# Load librariesimportnumpyasnpimportmatplotlib.pyplotaspltfromsklearn.ensembleimportRandomForestClassifierfromsklearnimportdatasets# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create random forest classifier objectrandomforest=RandomForestClassifier(random_state=0,n_jobs=-1)# Train modelmodel=randomforest.fit(features,target)# Calculate feature importancesimportances=model.feature_importances_# Sort feature importances in descending orderindices=np.argsort(importances)[::-1]# Rearrange feature names so they match the sorted feature importancesnames=[iris.feature_names[i]foriinindices]# Create plotplt.figure()# Create plot titleplt.title("Feature Importance")# Add barsplt.bar(range(features.shape[1]),importances[indices])# Add feature names as x-axis labelsplt.xticks(range(features.shape[1]),names,rotation=90)# Show plotplt.show()
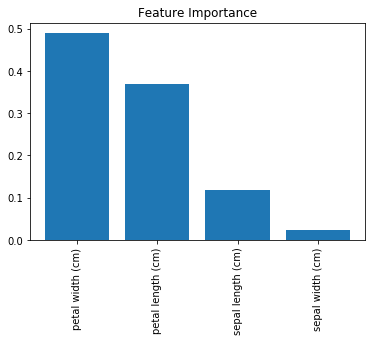
Discussion
One of the major benefits of decision trees is interpretability. Specifically, we can visualize the entire model (see Recipe 13.3). However, a random forest model is comprised of tens, hundreds, even thousands of decision trees. This makes a simple, intuitive visualization of a random forest model impractical. That said, there is another option: we can compare (and visualize) the relative importance of each feature.
In Recipe 13.3, we visualized a decision tree classifier model and saw that decision rules based only on petal width were able to classify many observations correctly. Intuitively, we can say that this means that petal width is an important feature in our classifier. More formally, features with splits that have the greater mean decrease in impurity (e.g., Gini impurity or entropy in classifiers and variance in regressors) are considered more important.
However, there are two things to keep in mind regarding feature importance. First, scikit-learn requires that we break up nominal categorical features into multiple binary features. This has the effect of spreading the importance of that feature across all of the binary features and can often make each feature appear to be unimportant even when the original nominal categorical feature is highly important. Second, if two features are highly correlated, one feature will claim much of the importance, making the other feature appear to be far less important—which has implications for interpretation if not considered.
In scikit-learn, classification and regression decision trees and random
forests can report the relative importance of each feature using the
feature_importances_ method:
# View feature importancesmodel.feature_importances_
array([ 0.11896532, 0.0231668 , 0.36804744, 0.48982043])
The higher the number, the more important the feature (all importance scores sum to 1). By plotting these values we can add interpretability to our random forest models.
14.7 Selecting Important Features in Random Forests
Solution
Identify the importance features and retrain the model using only the most important features:
# Load librariesfromsklearn.ensembleimportRandomForestClassifierfromsklearnimportdatasetsfromsklearn.feature_selectionimportSelectFromModel# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create random forest classifierrandomforest=RandomForestClassifier(random_state=0,n_jobs=-1)# Create object that selects features with importance greater# than or equal to a thresholdselector=SelectFromModel(randomforest,threshold=0.3)# Feature new feature matrix using selectorfeatures_important=selector.fit_transform(features,target)# Train random forest using most important featresmodel=randomforest.fit(features_important,target)
Discussion
There are situations where we might want to reduce the number of features in our model. For example, we might want to reduce the model’s variance or we might want to improve interpretability by including only the most important features.
In scikit-learn we can use a simple two-stage workflow to create a model
with reduced features. First, we train a random forest model using all
features. Then, we use this model to identify the most important
features. Next, we create a new feature matrix that includes only these
features. In our solution, we used the SelectFromModel method to create
a feature matrix containing only features with an importance greater
than or equal to some threshold value. Finally, we created a new model using
only those features.
It must be noted that there are two caveats to this approach. First, nominal categorical features that have been one-hot encoded will see the feature importance diluted across the binary features. Second, the feature importance of highly correlated features will be effectively assigned to one feature and not evenly distributed across both features.
14.8 Handling Imbalanced Classes
Solution
Train a decision tree or random forest model with
class_weight="balanced":
# Load librariesimportnumpyasnpfromsklearn.ensembleimportRandomForestClassifierfromsklearnimportdatasets# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Make class highly imbalanced by removing first 40 observationsfeatures=features[40:,:]target=target[40:]# Create target vector indicating if class 0, otherwise 1target=np.where((target==0),0,1)# Create random forest classifier objectrandomforest=RandomForestClassifier(random_state=0,n_jobs=-1,class_weight="balanced")# Train modelmodel=randomforest.fit(features,target)
Discussion
Imbalanced classes are a common problem when we are doing machine learning in the real world. Left unaddressed, the presence of imbalanced classes can reduce the performance of our model. We already talked about a few strategies for handling imbalanced classes during preprocessing in
Recipe 17.5. However, many learning algorithms in scikit-learn come with built-in methods for correcting for imbalanced classes. We can set RandomForestClassifier to correct for imbalanced classes using the class_weight parameter. If supplied with a dictionary in the form of class names and respective desired weights (e.g., {"male": 0.2, "female": 0.8}), RandomForestClassifier will weight the classes accordingly. However, often a more useful argument is
balanced, wherein classes are automatically weighted inversely
proportional to how frequently they appear in the data:
where wj is the weight to class j,
n is the number of observations, nj is the
number of observations in class j, and k is the total number of classes. For example, in our solution we have 2 classes (k), 110 observations (n), and 10 and 100 observations in each class, respectively (nj). If we weight the classes using class_weight="balanced", then the smaller
class is weighted more:
# Calculate weight for small class110/(2*10)
5.5
while the larger class is weighted less:
# Calculate weight for large class110/(2*100)
0.55
14.9 Controlling Tree Size
Solution
Use the tree structure parameters in scikit-learn tree-based learning algorithms:
# Load librariesfromsklearn.treeimportDecisionTreeClassifierfromsklearnimportdatasets# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create decision tree classifier objectdecisiontree=DecisionTreeClassifier(random_state=0,max_depth=None,min_samples_split=2,min_samples_leaf=1,min_weight_fraction_leaf=0,max_leaf_nodes=None,min_impurity_decrease=0)# Train modelmodel=decisiontree.fit(features,target)
Discussion
scikit-learn’s tree-based learning algorithms have a variety of techniques for controlling the size of the decision tree(s). These are accessed through parameters:
max_depth-
Maximum depth of the tree. If
None, the tree is grown until all leaves are pure. If an integer, the tree is effectively “pruned” to that depth. min_samples_split-
Minimum number of observations at a node before that node is split. If an integer is supplied as an argument it determines the raw minimum, while if a float is supplied the minimum is the percent of total observations.
min_samples_leaf-
Minimum number of observations required to be at a leaf. Uses the same arguments as
min_samples_split. max_leaf_nodes-
Maximum number of leaves.
min_impurity_split-
Minimum impurity decrease required before a split is performed.
While it is useful to know these parameters exist, most likely we will
only be using max_depth and min_impurity_split because shallower
trees (sometimes called stumps) are simpler models and thus have lower
variance.
14.10 Improving Performance Through Boosting
Solution
Train a boosted model using AdaBoostClassifier or AdaBoostRegressor:
# Load librariesfromsklearn.ensembleimportAdaBoostClassifierfromsklearnimportdatasets# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create adaboost tree classifier objectadaboost=AdaBoostClassifier(random_state=0)# Train modelmodel=adaboost.fit(features,target)
Discussion
In random forest, an ensemble (group) of randomized decision trees predicts the target vector. An alternative, and often more powerful, approach is called boosting. In one form of boosting called AdaBoost, we iteratively train a series of weak models (most often a shallow decision tree, sometimes called a stump), each iteration giving higher priority to observations the previous model predicted incorrectly. More specifically, in AdaBoost:
-
Assign every observation, xi, an initial weight value, , where n is the total number of observations in the data.
-
Train a “weak” model on the data.
-
For each observation:
-
If weak model predicts xi correctly, wi is increased.
-
If weak model predicts xi incorrectly, wi is decreased.
-
-
Train a new weak model where observations with greater wi are given greater priority.
-
Repeat steps 4 and 5 until the data is perfectly predicted or a preset number of weak models has been trained.
The end result is an aggregated model where individual weak models focus
on more difficult (from a prediction perspective) observations. In
scikit-learn, we can implement AdaBoost using AdaBoostClassifier or
AdaBoostRegressor. The most important parameters are base_estimator, n_estimators, and learning_rate:
-
base_estimatoris the learning algorithm to use to train the weak models. This will almost always not need to be changed because by far the most common learner to use with AdaBoost is a decision tree—the parameter’s default argument. -
n_estimatorsis the number of models to iteratively train. -
learning_rateis the contribution of each model to the weights and defaults to1. Reducing the learning rate will mean the weights will be increased or decreased to a small degree, forcing the model to train slower (but sometimes resulting in better performance scores). -
lossis exclusive toAdaBoostRegressorand sets the loss function to use when updating weights. This defaults to a linear loss function, but can be changed tosquareorexponential.
14.11 Evaluating Random Forests with Out-of-Bag Errors
Solution
Calculate the model’s out-of-bag score:
# Load librariesfromsklearn.ensembleimportRandomForestClassifierfromsklearnimportdatasets# Load datairis=datasets.load_iris()features=iris.datatarget=iris.target# Create random tree classifier objectrandomforest=RandomForestClassifier(random_state=0,n_estimators=1000,oob_score=True,n_jobs=-1)# Train modelmodel=randomforest.fit(features,target)# View out-of-bag-errorrandomforest.oob_score_
0.95333333333333337
Discussion
In random forests, each decision tree is trained using a bootstrapped subset of observations. This means that for every tree there is a separate subset of observations not being used to train that tree. These are called out-of-bag (OOB) observations. We can use OOB observations as a test set to evaluate the performance of our random forest.
For every observation, the learning algorithm compares the observation’s true value with the prediction from a subset of trees not trained using that observation. The overall score is calculated and provides a single measure of a random forest’s performance. OOB score estimation is an alternative to cross-validation.
In scikit-learn, we can OOB scores of a random forest by setting
oob_score=True in the random forest object (i.e.,
RandomForestClassifier). The score can be retrieved using
oob_score_.
